Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output
- PMID: 21738444
- PMCID: PMC3125158
- DOI: 10.1371/journal.pbio.1001086
Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output
Abstract
The basic unit of genome packaging is the nucleosome, and nucleosomes have long been proposed to restrict DNA accessibility both to damage and to transcription. Nucleosome number in cells was considered fixed, but recently aging yeast and mammalian cells were shown to contain fewer nucleosomes. We show here that mammalian cells lacking High Mobility Group Box 1 protein (HMGB1) contain a reduced amount of core, linker, and variant histones, and a correspondingly reduced number of nucleosomes, possibly because HMGB1 facilitates nucleosome assembly. Yeast nhp6 mutants lacking Nhp6a and -b proteins, which are related to HMGB1, also have a reduced amount of histones and fewer nucleosomes. Nucleosome limitation in both mammalian and yeast cells increases the sensitivity of DNA to damage, increases transcription globally, and affects the relative expression of about 10% of genes. In yeast nhp6 cells the loss of more than one nucleosome in four does not affect the location of nucleosomes and their spacing, but nucleosomal occupancy. The decrease in nucleosomal occupancy is non-uniform and can be modelled assuming that different nucleosomal sites compete for available histones. Sites with a high propensity to occupation are almost always packaged into nucleosomes both in wild type and nucleosome-depleted cells; nucleosomes on sites with low propensity to occupation are disproportionately lost in nucleosome-depleted cells. We suggest that variation in nucleosome number, by affecting nucleosomal occupancy both genomewide and gene-specifically, constitutes a novel layer of epigenetic regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


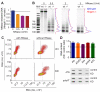


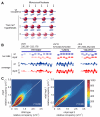
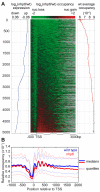
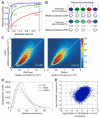
Comment in
-
Not all genes are equal; shortage of histones affects some genes more than others.PLoS Biol. 2011 Jun;9(6):e1001098. doi: 10.1371/journal.pbio.1001098. Epub 2011 Jun 28. PLoS Biol. 2011. PMID: 21738447 Free PMC article. No abstract available.
References
-
- Kornberg R. D, Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. - PubMed
-
- Li B, Carey M, Workman J. L. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
-
- Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. - PubMed
-
- van Holde K. E. Chromatin; In: Rich A, editor. New York: Springer-Verlag; 1988. 497
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

