Network clustering revealed the systemic alterations of mitochondrial protein expression
- PMID: 21738461
- PMCID: PMC3127811
- DOI: 10.1371/journal.pcbi.1002093
Network clustering revealed the systemic alterations of mitochondrial protein expression
Abstract
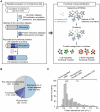
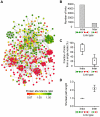
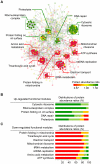
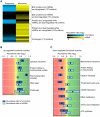
The mitochondrial protein repertoire varies depending on the cellular state. Protein component modifications caused by mitochondrial DNA (mtDNA) depletion are related to a wide range of human diseases; however, little is known about how nuclear-encoded mitochondrial proteins (mt proteome) changes under such dysfunctional states. In this study, we investigated the systemic alterations of mtDNA-depleted (ρ(0)) mitochondria by using network analysis of gene expression data. By modularizing the quantified proteomics data into protein functional networks, systemic properties of mitochondrial dysfunction were analyzed. We discovered that up-regulated and down-regulated proteins were organized into two predominant subnetworks that exhibited distinct biological processes. The down-regulated network modules are involved in typical mitochondrial functions, while up-regulated proteins are responsible for mtDNA repair and regulation of mt protein expression and transport. Furthermore, comparisons of proteome and transcriptome data revealed that ρ(0) cells attempted to compensate for mtDNA depletion by modulating the coordinated expression/transport of mt proteins. Our results demonstrate that mt protein composition changed to remodel the functional organization of mitochondrial protein networks in response to dysfunctional cellular states. Human mt protein functional networks provide a framework for understanding how cells respond to mitochondrial dysfunctions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

