The SV40 late protein VP4 is a viroporin that forms pores to disrupt membranes for viral release
- PMID: 21738474
- PMCID: PMC3128117
- DOI: 10.1371/journal.ppat.1002116
The SV40 late protein VP4 is a viroporin that forms pores to disrupt membranes for viral release
Abstract
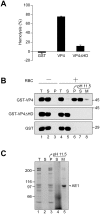
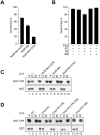
Nonenveloped viruses are generally released by the timely lysis of the host cell by a poorly understood process. For the nonenveloped virus SV40, virions assemble in the nucleus and then must be released from the host cell without being encapsulated by cellular membranes. This process appears to involve the well-controlled insertion of viral proteins into host cellular membranes rendering them permeable to large molecules. VP4 is a newly identified SV40 gene product that is expressed at late times during the viral life cycle that corresponds to the time of cell lysis. To investigate the role of this late expressed protein in viral release, water-soluble VP4 was expressed and purified as a GST fusion protein from bacteria. Purified VP4 was found to efficiently bind biological membranes and support their disruption. VP4 perforated membranes by directly interacting with the membrane bilayer as demonstrated by flotation assays and the release of fluorescent markers encapsulated into large unilamellar vesicles or liposomes. The central hydrophobic domain of VP4 was essential for membrane binding and disruption. VP4 displayed a preference for membranes comprised of lipids that replicated the composition of the plasma membranes over that of nuclear membranes. Phosphatidylethanolamine, a lipid found at high levels in bacterial membranes, was inhibitory against the membrane perforation activity of VP4. The disruption of membranes by VP4 involved the formation of pores of ∼3 nm inner diameter in mammalian cells including permissive SV40 host cells. Altogether, these results support a central role of VP4 acting as a viroporin in the perforation of cellular membranes to trigger SV40 viral release.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Pornillos O, Garrus JE, Sundquist WI. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 2002;12:569–579. - PubMed
-
- Han Z, Harty RN. The NS3 protein of bluetongue virus exhibits viroporin-like properties. J Biol Chem. 2004;279:43092–43097. - PubMed
-
- Aldabe R, Barco A, Carrasco L. Membrane permeabilization by poliovirus proteins 2B and 2BC. J Biol Chem. 1996;271:23134–23137. - PubMed
-
- Fiers W, Contreras R, Haegemann G, Rogiers R, Van de Voorde A, et al. Complete nucleotide sequence of SV40 DNA. Nature. 1978;273:113–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

