Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: maintenance of a reservoir for genetic plasticity
- PMID: 21738490
- PMCID: PMC3128108
- DOI: 10.1371/journal.pgen.1002156
Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: maintenance of a reservoir for genetic plasticity
Abstract
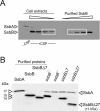
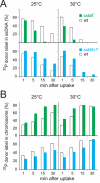
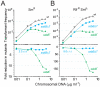
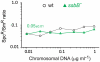
Bacteria encode a single-stranded DNA (ssDNA) binding protein (SSB) crucial for genome maintenance. In Bacillus subtilis and Streptococcus pneumoniae, an alternative SSB, SsbB, is expressed uniquely during competence for genetic transformation, but its precise role has been disappointingly obscure. Here, we report our investigations involving comparison of a null mutant (ssbB(-)) and a C-ter truncation (ssbBΔ7) of SsbB of S. pneumoniae, the latter constructed because SSBs' acidic tail has emerged as a key site for interactions with partner proteins. We provide evidence that SsbB directly protects internalized ssDNA. We show that SsbB is highly abundant, potentially allowing the binding of ~1.15 Mb ssDNA (half a genome equivalent); that it participates in the processing of ssDNA into recombinants; and that, at high DNA concentration, it is of crucial importance for chromosomal transformation whilst antagonizing plasmid transformation. While the latter observation explains a long-standing observation that plasmid transformation is very inefficient in S. pneumoniae (compared to chromosomal transformation), the former supports our previous suggestion that SsbB creates a reservoir of ssDNA, allowing successive recombination cycles. SsbBΔ7 fulfils the reservoir function, suggesting that SsbB C-ter is not necessary for processing protein(s) to access stored ssDNA. We propose that the evolutionary raison d'être of SsbB and its abundance is maintenance of this reservoir, which contributes to the genetic plasticity of S. pneumoniae by increasing the likelihood of multiple transformation events in the same cell.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Hiller NL, Ahmed A, Powell E, Martin DP, Eutsey R, et al. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 2010;6:e1001108. doi: 10.1371/journal.ppat.1001108. - DOI - PMC - PubMed
-
- Lacks S, Greenberg B, Carlson K. Fate of donor DNA in pneumococcal transformation. J Mol Biol. 1967;29:327–347.
-
- Dagkessamanskaia A, Moscoso M, Hénard V, Guiral S, Overweg K, et al. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol Microbiol. 2004;51:1071–1086. - PubMed
-
- Peterson S, Sung CK, Cline R, Desai BV, Snesrud E, et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae. Mol Microbiol. 2004;51:1051–1070. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

