Application of MLST and pilus gene sequence comparisons to investigate the population structures of Actinomyces naeslundii and Actinomyces oris
- PMID: 21738661
- PMCID: PMC3127948
- DOI: 10.1371/journal.pone.0021430
Application of MLST and pilus gene sequence comparisons to investigate the population structures of Actinomyces naeslundii and Actinomyces oris
Abstract
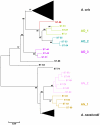
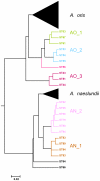
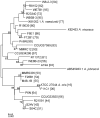
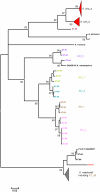
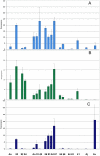
Actinomyces naeslundii and Actinomyces oris are members of the oral biofilm. Their identification using 16S rRNA sequencing is problematic and better achieved by comparison of metG partial sequences. A. oris is more abundant and more frequently isolated than A. naeslundii. We used a multi-locus sequence typing approach to investigate the genotypic diversity of these species and assigned A. naeslundii (n = 37) and A. oris (n = 68) isolates to 32 and 68 sequence types (ST), respectively. Neighbor-joining and ClonalFrame dendrograms derived from the concatenated partial sequences of 7 house-keeping genes identified at least 4 significant subclusters within A. oris and 3 within A. naeslundii. The strain collection we had investigated was an under-representation of the total population since at least 3 STs composed of single strains may represent discrete clusters of strains not well represented in the collection. The integrity of these sub-clusters was supported by the sequence analysis of fimP and fimA, genes coding for the type 1 and 2 fimbriae, respectively. An A. naeslundii subcluster was identified with both fimA and fimP genes and these strains were able to bind to MUC7 and statherin while all other A. naeslundii strains possessed only fimA and did not bind to statherin. An A. oris subcluster harboured a fimA gene similar to that of Actinomyces odontolyticus but no detectable fimP failed to bind significantly to either MUC7 or statherin. These data are evidence of extensive genotypic and phenotypic diversity within the species A. oris and A. naeslundii but the status of the subclusters identified here will require genome comparisons before their phylogenic position can be unequivocally established.
Conflict of interest statement
Figures

References
-
- Bowden GH, Nolette N, Ryding H, Cleghorn BM. The diversity and distribution of the predominant ribotypes of Actinomyces naeslundii genospecies 1 and 2 in samples from enamel and from healthy and carious root surfaces of teeth. J Dent Res. 1999;78:1800–1809. - PubMed
-
- Henssge U, Do T, Radford DR, Gilbert SC, Clark D, et al. Emended description of Actinomyces naelsundii and descriptions of Actinomyces oris sp. nov. and Actinomyces johnsonii sp. nov., previously identified as Actinomyces naeslundii genospecies 1, 2 and WVA 963. Int J Syst Evol Microbiol. 2009;59:509–516. - PMC - PubMed
-
- Li J, Helmerhorst EJ, Leone CW, Troxler RF, Yaskell T, et al. Identification of early microbial colonizers in human dental biofilm. J Appl Microbiol. 2004;97:1311–1318. - PubMed
-
- Johnson JL, Moore LV, Kaneko B, Moore WE. Actinomyces georgiae sp. nov., Actinomyces gerencseriae sp. nov., designation of two genospecies of Actinomyces naeslundii, and inclusion of A. naeslundii serotypes II and III and Actinomyces viscosus serotype II in A. naeslundii genospecies 2. Int J Syst Bacteriol. 1990;40:273–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

