Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors
- PMID: 21738689
- PMCID: PMC3124521
- DOI: 10.1371/journal.pone.0021524
Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors
Abstract
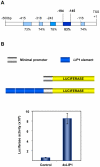
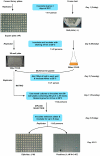
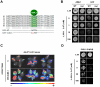
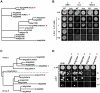
Transcriptional regulation is an important mechanism underlying gene expression and has played a crucial role in evolution. The number, position and interactions between cis-elements and transcription factors (TFs) determine the expression pattern of a gene. To identify functionally relevant cis-elements in gene promoters, a phylogenetic shadowing approach with a lipase gene (LIP1) was used. As a proof of concept, in silico analyses of several Brassicaceae LIP1 promoters identified a highly conserved sequence (LIP1 element) that is sufficient to drive strong expression of a reporter gene in planta. A collection of ca. 1,200 Arabidopsis thaliana TF open reading frames (ORFs) was arrayed in a 96-well format (RR library) and a convenient mating based yeast one hybrid (Y1H) screening procedure was established. We constructed an episomal plasmid (pTUY1H) to clone the LIP1 element and used it as bait for Y1H screenings. A novel interaction with an HD-ZIP (AtML1) TF was identified and abolished by a 2 bp mutation in the LIP1 element. A role of this interaction in transcriptional regulation was confirmed in planta. In addition, we validated our strategy by reproducing the previously reported interaction between a MYB-CC (PHR1) TF, a central regulator of phosphate starvation responses, with a conserved promoter fragment (IPS1 element) containing its cognate binding sequence. Finally, we established that the LIP1 and IPS1 elements were differentially bound by HD-ZIP and MYB-CC family members in agreement with their genetic redundancy in planta. In conclusion, combining in silico analyses of orthologous gene promoters with Y1H screening of the RR library represents a powerful approach to decipher cis- and trans-regulatory codes.
Conflict of interest statement
Figures

References
-
- Singh K, Foley RC, Oñate-Sánchez L. Transcription factors in plant defense and stress responses. Curr Opin Plant Biol. 2002;5:430–436. - PubMed
-
- Cliften PF, Hillier LW, Fulton L, Graves T, Miner T, et al. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 2001;11:1175–86. - PubMed
-
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

