Enzymes are enriched in bacterial essential genes
- PMID: 21738765
- PMCID: PMC3125301
- DOI: 10.1371/journal.pone.0021683
Enzymes are enriched in bacterial essential genes
Abstract
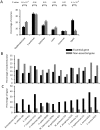
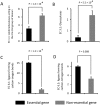
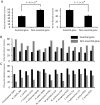
Essential genes, those indispensable for the survival of an organism, play a key role in the emerging field, synthetic biology. Characterization of functions encoded by essential genes not only has important practical implications, such as in identifying antibiotic drug targets, but can also enhance our understanding of basic biology, such as functions needed to support cellular life. Enzymes are critical for almost all cellular activities. However, essential genes have not been systematically examined from the aspect of enzymes and the chemical reactions that they catalyze. Here, by comprehensively analyzing essential genes in 14 bacterial genomes in which large-scale gene essentiality screens have been performed, we found that enzymes are enriched in essential genes. Essential enzymes have overrepresented ligases (especially those forming carbon-oxygen bonds and carbon-nitrogen bonds), nucleotidyltransferases and phosphotransferases, while have underrepresented oxidoreductases. Furthermore, essential enzymes tend to associate with more gene ontology domains. These results, from the aspect of chemical reactions, provide further insights into the understanding of functions needed to support natural cellular life, as well as synthetic cells, and provide additional parameters that can be integrated into gene essentiality prediction algorithms.
Conflict of interest statement
Figures

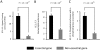

Similar articles
-
Protein localization analysis of essential genes in prokaryotes.Sci Rep. 2014 Aug 8;4:6001. doi: 10.1038/srep06001. Sci Rep. 2014. PMID: 25105358 Free PMC article.
-
Machine learning approach to gene essentiality prediction: a review.Brief Bioinform. 2021 Sep 2;22(5):bbab128. doi: 10.1093/bib/bbab128. Brief Bioinform. 2021. PMID: 33842944 Review.
-
Essential bacterial functions encoded by gene pairs.J Bacteriol. 2007 Jan;189(2):591-602. doi: 10.1128/JB.01381-06. Epub 2006 Nov 17. J Bacteriol. 2007. PMID: 17114254 Free PMC article.
-
Gene essentiality analysis based on DEG, a database of essential genes.Methods Mol Biol. 2008;416:391-400. doi: 10.1007/978-1-59745-321-9_27. Methods Mol Biol. 2008. PMID: 18392983
-
Essential genes on metabolic maps.Curr Opin Biotechnol. 2006 Oct;17(5):448-56. doi: 10.1016/j.copbio.2006.08.006. Epub 2006 Sep 15. Curr Opin Biotechnol. 2006. PMID: 16978855 Review.
Cited by
-
Exploring the enzymatic repertoires of Bacteria and Archaea and their associations with metabolic maps.Braz J Microbiol. 2024 Dec;55(4):3147-3157. doi: 10.1007/s42770-024-01462-3. Epub 2024 Jul 25. Braz J Microbiol. 2024. PMID: 39052173 Free PMC article.
-
Recent advances in the characterization of essential genes and development of a database of essential genes.Imeta. 2024 Jan 2;3(1):e157. doi: 10.1002/imt2.157. eCollection 2024 Feb. Imeta. 2024. PMID: 38868518 Free PMC article. Review.
-
Comparative analysis of the Burkholderia cenocepacia K56-2 essential genome reveals cell envelope functions that are uniquely required for survival in species of the genus Burkholderia.Microb Genom. 2017 Nov;3(11):e000140. doi: 10.1099/mgen.0.000140. Microb Genom. 2017. PMID: 29208119 Free PMC article.
-
Cas9 Nickase-Assisted RNA Repression Enables Stable and Efficient Manipulation of Essential Metabolic Genes in Clostridium cellulolyticum.Front Microbiol. 2017 Sep 7;8:1744. doi: 10.3389/fmicb.2017.01744. eCollection 2017. Front Microbiol. 2017. PMID: 28936208 Free PMC article.
-
Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates.PLoS One. 2012;7(8):e43080. doi: 10.1371/journal.pone.0043080. Epub 2012 Aug 13. PLoS One. 2012. PMID: 22912793 Free PMC article.
References
-
- Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, et al. Genome transplantation in bacteria: changing one species to another. Science. 2007;317:632–638. - PubMed
-
- Henkel J, Maurer SM. Parts, property and sharing. Nat Biotechnol. 2009;27:1095–1098. - PubMed
-
- de S Cameron NM, Caplan A. Our synthetic future. Nat Biotechnol. 2009;27:1103–1105. - PubMed
-
- May M. Engineering a new business. Nat Biotechnol. 2009;27:1112–1120. - PubMed
-
- Pennisi E. Genomics. Synthetic genome brings new life to bacterium. Science. 2010;328:958–959. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

