A polymorphism in the splice donor site of ZNF419 results in the novel renal cell carcinoma-associated minor histocompatibility antigen ZAPHIR
- PMID: 21738768
- PMCID: PMC3125305
- DOI: 10.1371/journal.pone.0021699
A polymorphism in the splice donor site of ZNF419 results in the novel renal cell carcinoma-associated minor histocompatibility antigen ZAPHIR
Abstract
Nonmyeloablative allogeneic stem cell transplantation (SCT) can induce remission in patients with renal cell carcinoma (RCC), but this graft-versus-tumor (GVT) effect is often accompanied by graft-versus-host disease (GVHD). Here, we evaluated minor histocompatibility antigen (MiHA)-specific T cell responses in two patients with metastatic RCC who were treated with reduced-intensity conditioning SCT followed by donor lymphocyte infusion (DLI). One patient had stable disease and emergence of SMCY.A2-specific CD8+ T cells was observed after DLI with the potential of targeting SMCY-expressing RCC tumor cells. The second patient experienced partial regression of lung metastases from whom we isolated a MiHA-specific CTL clone with the capability of targeting RCC cell lines. Whole genome association scanning revealed that this CTL recognizes a novel HLA-B7-restricted MiHA, designated ZAPHIR, resulting from a polymorphism in the splice donor site of the ZNF419 gene. Tetramer analysis showed that emergence of ZAPHIR-specific CD8+ T cells in peripheral blood occurred in the absence of GVHD. Furthermore, the expression of ZAPHIR in solid tumor cell lines indicates the involvement of ZAPHIR-specific CD8+ T cell responses in selective GVT immunity. These findings illustrate that the ZNF419-encoded MiHA ZAPHIR is an attractive target for specific immunotherapy after allogeneic SCT.
Conflict of interest statement
Figures



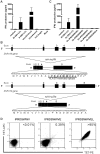
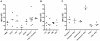

References
-
- Yang JC, Childs R. Immunotherapy for renal cell cancer. J Clin Oncol. 2006;24:5576–5583. - PubMed
-
- Childs R, Chernoff A, Contentin N, Bahceci E, Schrump D, et al. Regression of metastatic renal-cell carcinoma after nonmyeloablative allogeneic peripheral-blood stem-cell transplantation. N Engl J Med. 2000;343:750–758. - PubMed
-
- Pedrazzoli P, Da Prada GA, Giorgiani G, Schiavo R, Zambelli A, et al. Allogeneic blood stem cell transplantation after a reduced-intensity, preparative regimen - A pilot study in patients with refractory malignancies. Cancer. 2002;94:2409–2415. - PubMed
-
- Rini BI, Zimmerman T, Stadler WM, Gajewski TF, Vogelzang NJ. Allogeneic stem-cell transplantation of renal cell cancer after nonmyeloablative chemotherapy: feasibility, engraftment, and clinical results. J Clin Oncol. 2002;20:2017–2024. - PubMed
-
- Tykodi SS, Warren EH, Thompson JA, Riddell SR, Childs RW, et al. Allogeneic hematopoietic cell transplantation for metastatic renal cell carcinoma after nonmyeloablative conditioning: toxicity, clinical response, and immunological response to minor histocompatibility antigens. Clin Cancer Res. 2004;10:7799–7811. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

