H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G₁ phase
- PMID: 21738837
- PMCID: PMC3127096
- DOI: 10.4161/nucl.2.2.15211
H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G₁ phase
Abstract
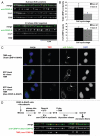
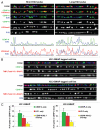
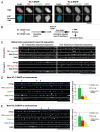
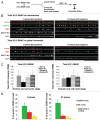
Centromeres are key regions of eukaryotic chromosomes that ensure proper chromosome segregation at cell division. In most eukaryotes, centromere identity is defined epigenetically by the presence of a centromeric histone H3 variant CenH3, called CENP-A in humans. How CENP-A is incorporated and reproducibly transmitted during the cell cycle is at the heart of this fundamental epigenetic mechanism. Centromeric DNA is replicated during S phase; however unlike replication-coupled assembly of canonical histones during S phase, newly synthesized CENP-A deposition at centromeres is restricted to a discrete time in late telophase/early G(1). These observations raise an important question: when 'old' CENP-A nucleosomes are segregated at the replication fork, are the resulting 'gaps' maintained until the next G(1), or are they filled by H3 nucleosomes during S phase and replaced by CENP-A in the following G(1)? Understanding such molecular mechanisms is important to reveal the composition/organization of centromeres in mitosis, when the kinetochore forms and functions. Here we investigate centromeric chromatin status during the cell cycle, using the SNAP-tag methodology to visualize old and new histones on extended chromatin fibers in human cells. Our results show that (1) both histone H3 variants H3.1 and H3.3 are deposited at centromeric domains in S phase and (2) there is reduced H3.3 (but not reduced H3.1) at centromeres in G(1) phase compared to S phase. These observations are consistent with a replacement model, where both H3.1 and H3.3 are deposited at centromeres in S phase and 'placeholder' H3.3 is replaced with CENP-A in G(1).
Keywords: CENP-A; DNA replication; cell cycle; centromere; histone deposition; kinetochore; mitosis.
Figures
References
-
- Sullivan KF. A solid foundation: Functional specialization of centromeric chromatin. Curr Opin Genet Dev. 2001;11:182–188. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
