Molecular determinants of disease in coxsackievirus B1 murine infection
- PMID: 21739448
- PMCID: PMC3677187
- DOI: 10.1002/jmv.22133
Molecular determinants of disease in coxsackievirus B1 murine infection
Abstract
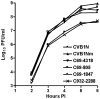
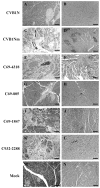
To understand better how different genomic regions may confer pathogenicity for the coxsackievirus B (CVB), two intratypic CVB1 variants, and a number of recombinant viruses were studied. Sequencing analysis showed 23 nucleotide changes between the parental non-pathogenic CVB1N and the pathogenic CVB1Nm. Mutations present in CVB1Nm were more conserved than those in CVB1N when compared to other CVB sequences. Inoculation in C3H/HeJ mice showed that the P1 region is critical for pathogenicity in murine pancreas and heart. The molecular determinants of disease for these organs partially overlap. Several P1 region amino acid differences appear to be located in the decay-accelerating factor (DAF) footprint CVBs. CVB1N and CVB1Nm interacted with human CAR, but only CVB1N seemed to interact with human DAF, as determined using soluble receptors in a plaque-reduction assay. However, the murine homolog Daf-1 did not interact with any virus assessed by hemagglutination. The results of this study suggest that an unknown receptor interaction with the virus play an important role in the pathogenicity of CVB1Nm. Further in vivo studies may clarify this issue.
Copyright © 2011 Wiley-Liss, Inc.
Figures



References
-
- Beck MA, Levander OA, Handy J. Selenium deficiency and viral infection. J Nutr. 2003;133:1463S–1467S. - PubMed
-
- Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

