Thermodynamics and kinetics of the hairpin ribozyme from atomistic folding/unfolding simulations
- PMID: 21740912
- PMCID: PMC3508787
- DOI: 10.1016/j.jmb.2011.06.042
Thermodynamics and kinetics of the hairpin ribozyme from atomistic folding/unfolding simulations
Abstract
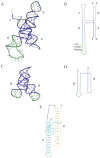
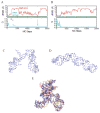
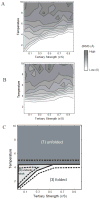
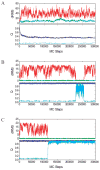
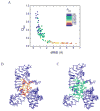
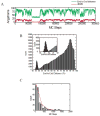
We report a set of atomistic folding/unfolding simulations for the hairpin ribozyme using a Monte Carlo algorithm. The hairpin ribozyme folds in solution and catalyzes self-cleavage or ligation via a specific two-domain structure. The minimal active ribozyme has been studied extensively, showing stabilization of the active structure by cations and dynamic motion of the active structure. Here, we introduce a simple model of tertiary-structure formation that leads to a phase diagram for the RNA as a function of temperature and tertiary-structure strength. We then employ this model to capture many folding/unfolding events and to examine the transition-state ensemble (TSE) of the RNA during folding to its active "docked" conformation. The TSE is compact but with few tertiary interactions formed, in agreement with single-molecule dynamics experiments. To compare with experimental kinetic parameters, we introduce a novel method to benchmark Monte Carlo kinetic parameters to docking/undocking rates collected over many single molecular trajectories. We find that topology alone, as encoded in a biased potential that discriminates between secondary and tertiary interactions, is sufficient to predict the thermodynamic behavior and kinetic folding pathway of the hairpin ribozyme. This method should be useful in predicting folding transition states for many natural or man-made RNA tertiary structures.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


Similar articles
-
Thermodynamics and kinetics of RNA tertiary structure formation in the junctionless hairpin ribozyme.Biophys Chem. 2017 Sep;228:62-68. doi: 10.1016/j.bpc.2017.07.001. Epub 2017 Jul 8. Biophys Chem. 2017. PMID: 28710920 Free PMC article.
-
The tertiary structure of the hairpin ribozyme is formed through a slow conformational search.Biochemistry. 2005 Mar 29;44(12):4870-6. doi: 10.1021/bi047772i. Biochemistry. 2005. PMID: 15779913
-
Characterization of a local folding event of the Tetrahymena group I ribozyme: effects of oligonucleotide substrate length, pH, and temperature on the two substrate binding steps.Biochemistry. 1999 Oct 26;38(43):14192-204. doi: 10.1021/bi9914309. Biochemistry. 1999. PMID: 10571993
-
RNA folding and the origins of catalytic activity in the hairpin ribozyme.Blood Cells Mol Dis. 2007 Jan-Feb;38(1):8-14. doi: 10.1016/j.bcmd.2006.10.004. Epub 2006 Dec 4. Blood Cells Mol Dis. 2007. PMID: 17150385 Review.
-
Structure and function of the hairpin ribozyme.J Mol Biol. 2000 Mar 24;297(2):269-91. doi: 10.1006/jmbi.2000.3560. J Mol Biol. 2000. PMID: 10715200 Review.
Cited by
-
A coarse-grained protein model in a water-like solvent.Sci Rep. 2013;3:1841. doi: 10.1038/srep01841. Sci Rep. 2013. PMID: 23674146 Free PMC article.
-
Thermodynamics and kinetics of RNA tertiary structure formation in the junctionless hairpin ribozyme.Biophys Chem. 2017 Sep;228:62-68. doi: 10.1016/j.bpc.2017.07.001. Epub 2017 Jul 8. Biophys Chem. 2017. PMID: 28710920 Free PMC article.
References
-
- Lilley DM. Structure, folding and catalysis of the small nucleolytic ribozymes. Current opinion in structural biology. 1999;9:330–8. - PubMed
-
- Lilley DM. Structure, folding and mechanisms of ribozymes. Current opinion in structural biology. 2005;15:313–23. - PubMed
-
- Hampel A, Tritz R. RNA catalytic properties of the minimum (−)sTRSV sequence. Biochemistry. 1989;28:4929–33. - PubMed
-
- Buzayan JM, Gerlach WL, Bruening G. Nonenzymatic Cleavage and Ligation of Rnas Complementary to a Plant-Virus Satellite Rna. Nature. 1986;323:349–353.
-
- Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–6. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

