Evaluation of FKBP and DHFR based destabilizing domains in Saccharomyces cerevisiae
- PMID: 21741238
- PMCID: PMC3156383
- DOI: 10.1016/j.bmcl.2011.06.006
Evaluation of FKBP and DHFR based destabilizing domains in Saccharomyces cerevisiae
Abstract
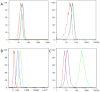
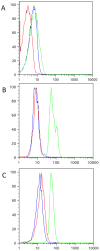
Two orthogonal destabilizing domains have been developed based on mutants of human FKBP12 as well as bacterial DHFR and these engineered domains have been used to control protein concentration in a variety of contexts in vitro and in vivo. FKBP12 based destabilizing domains cannot be rescued in the yeast Saccharomyces cerevisiae; ecDHFR based destabilizing domains are not degraded as efficiently in S. cerevisiae as in mammalian cells or Plasmodium, but provide a starting point for the development of domains with increased signal-to-noise in S. cerevisiae.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
The rapamycin-binding domain of the protein kinase mammalian target of rapamycin is a destabilizing domain.J Biol Chem. 2007 May 4;282(18):13395-401. doi: 10.1074/jbc.M700498200. Epub 2007 Mar 9. J Biol Chem. 2007. PMID: 17350953 Free PMC article.
-
A yeast sensor of ligand binding.Nat Biotechnol. 2001 Nov;19(11):1042-6. doi: 10.1038/nbt1101-1042. Nat Biotechnol. 2001. PMID: 11689849
-
Functions of FKBP12 and mitochondrial cyclophilin active site residues in vitro and in vivo in Saccharomyces cerevisiae.Mol Biol Cell. 1997 Nov;8(11):2267-80. doi: 10.1091/mbc.8.11.2267. Mol Biol Cell. 1997. PMID: 9362068 Free PMC article.
-
Prolyl isomerases in yeast.Front Biosci. 2004 Sep 1;9:2420-46. doi: 10.2741/1405. Front Biosci. 2004. PMID: 15353296 Review.
-
Physiological function of FKBP12, a primary target of rapamycin/FK506: a newly identified role in transcription of ribosomal protein genes in yeast.Curr Genet. 2021 Jun;67(3):383-388. doi: 10.1007/s00294-020-01142-3. Epub 2021 Jan 12. Curr Genet. 2021. PMID: 33438053 Review.
Cited by
-
Utility of the DHFR-based destabilizing domain across mouse models of retinal degeneration and aging.iScience. 2022 Apr 6;25(5):104206. doi: 10.1016/j.isci.2022.104206. eCollection 2022 May 20. iScience. 2022. PMID: 35521529 Free PMC article.
-
Destabilization domain approach adapted for regulated protein expression in the protozoan parasite Entamoeba histolytica.Int J Parasitol. 2014 Sep;44(10):729-35. doi: 10.1016/j.ijpara.2014.05.002. Epub 2014 Jun 11. Int J Parasitol. 2014. PMID: 24929134 Free PMC article.
-
Chemical biology strategies for posttranslational control of protein function.Chem Biol. 2014 Sep 18;21(9):1238-52. doi: 10.1016/j.chembiol.2014.08.011. Chem Biol. 2014. PMID: 25237866 Free PMC article. Review.
-
Deep mutational scanning of the RNase III-like domain in Trypanosoma brucei RNA editing protein KREPB4.Front Cell Infect Microbiol. 2024 Apr 8;14:1381155. doi: 10.3389/fcimb.2024.1381155. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38650737 Free PMC article.
-
A general strategy to construct small molecule biosensors in eukaryotes.Elife. 2015 Dec 29;4:e10606. doi: 10.7554/eLife.10606. Elife. 2015. PMID: 26714111 Free PMC article.
References
-
- Armstrong CM, Goldberg DE. Nature methods. 2007;4:1007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

