Targeting androgen receptor in estrogen receptor-negative breast cancer
- PMID: 21741601
- PMCID: PMC3180861
- DOI: 10.1016/j.ccr.2011.05.026
Targeting androgen receptor in estrogen receptor-negative breast cancer
Abstract
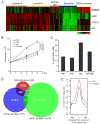
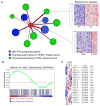
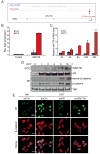
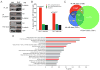
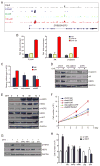
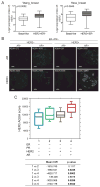
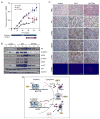
Endocrine therapies for breast cancer that target the estrogen receptor (ER) are ineffective in the 25%-30% of cases that are ER negative (ER-). Androgen receptor (AR) is expressed in 60%-70% of breast tumors, independent of ER status. How androgens and AR regulate breast cancer growth remains largely unknown. We find that AR is enriched in ER- breast tumors that overexpress HER2. Through analysis of the AR cistrome and androgen-regulated gene expression in ER-/HER2+ breast cancers we find that AR mediates ligand-dependent activation of Wnt and HER2 signaling pathways through direct transcriptional induction of WNT7B and HER3. Specific targeting of AR, Wnt or HER2 signaling impairs androgen-stimulated tumor cell growth suggesting potential therapeutic approaches for ER-/HER2+ breast cancers.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

References
-
- Agoff SN, Swanson PE, Linden H, Hawes SE, Lawton TJ. Androgen receptor expression in estrogen receptor-negative breast cancer. Immunohistochemical, clinical, and prognostic associations. Am J Clin Pathol. 2003;120:725–731. - PubMed
-
- Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. - PubMed
-
- Bonnefoi H, Potti A, Delorenzi M, Mauriac L, Campone M, Tubiana-Hulin M, Petit T, Rouanet P, Jassem J, Blot E, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8:1071–1078. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

