Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome
- PMID: 21741921
- PMCID: PMC3417828
- DOI: 10.1053/j.gastro.2011.06.072
Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome
Abstract
Background & aims: The intestinal microbiomes of healthy children and pediatric patients with irritable bowel syndrome (IBS) are not well defined. Studies in adults have indicated that the gastrointestinal microbiota could be involved in IBS.
Methods: We analyzed 71 samples from 22 children with IBS (pediatric Rome III criteria) and 22 healthy children, ages 7-12 years, by 16S ribosomal RNA gene sequencing, with an average of 54,287 reads/stool sample (average 454 read length = 503 bases). Data were analyzed using phylogenetic-based clustering (Unifrac), or an operational taxonomic unit (OTU) approach using a supervised machine learning tool (randomForest). Most samples were also hybridized to a microarray that can detect 8741 bacterial taxa (16S rRNA PhyloChip).
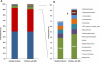
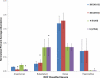
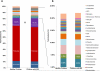
Results: Microbiomes associated with pediatric IBS were characterized by a significantly greater percentage of the class γ-proteobacteria (0.07% vs 0.89% of total bacteria, respectively; P < .05); 1 prominent component of this group was Haemophilus parainfluenzae. Differences highlighted by 454 sequencing were confirmed by high-resolution PhyloChip analysis. Using supervised learning techniques, we were able to classify different subtypes of IBS with a success rate of 98.5%, using limited sets of discriminant bacterial species. A novel Ruminococcus-like microbe was associated with IBS, indicating the potential utility of microbe discovery for gastrointestinal disorders. A greater frequency of pain correlated with an increased abundance of several bacterial taxa from the genus Alistipes.
Conclusions: Using 16S metagenomics by PhyloChip DNA hybridization and deep 454 pyrosequencing, we associated specific microbiome signatures with pediatric IBS. These findings indicate the important association between gastrointestinal microbes and IBS in children; these approaches might be used in diagnosis of functional bowel disorders in pediatric patients.
Copyright © 2011 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures
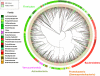
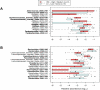

 IBS-C: IBS with constipation (n=41 samples),
IBS-C: IBS with constipation (n=41 samples),  IBS-U: unsubtyped IBS (n=22 samples). Bray-Curtis analysis was used to generate a matrix of pairwise sample dissimilarities between communities. The scatterplot was generated from the matrix of distances using principal components analysis. Data were generated by 454 pyrosequencing (V1–V3 region only, 2 replicates per sample).
IBS-U: unsubtyped IBS (n=22 samples). Bray-Curtis analysis was used to generate a matrix of pairwise sample dissimilarities between communities. The scatterplot was generated from the matrix of distances using principal components analysis. Data were generated by 454 pyrosequencing (V1–V3 region only, 2 replicates per sample).Comment in
-
Bugs, stool, and the irritable bowel syndrome: too much is as bad as too little?Gastroenterology. 2011 Nov;141(5):1555-9. doi: 10.1053/j.gastro.2011.09.019. Epub 2011 Sep 22. Gastroenterology. 2011. PMID: 21945058 No abstract available.
Similar articles
-
Leveraging Human Microbiome Features to Diagnose and Stratify Children with Irritable Bowel Syndrome.J Mol Diagn. 2019 May;21(3):449-461. doi: 10.1016/j.jmoldx.2019.01.006. Epub 2019 Apr 17. J Mol Diagn. 2019. PMID: 31005411 Free PMC article.
-
Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome.Gastroenterology. 2011 Nov;141(5):1792-801. doi: 10.1053/j.gastro.2011.07.043. Epub 2011 Aug 5. Gastroenterology. 2011. PMID: 21820992
-
Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome.Gastroenterology. 2017 Jan;152(1):111-123.e8. doi: 10.1053/j.gastro.2016.09.049. Epub 2016 Oct 7. Gastroenterology. 2017. PMID: 27725146
-
Gastrointestinal microbiota in irritable bowel syndrome: present state and perspectives.Microbiology (Reading). 2010 Nov;156(Pt 11):3205-3215. doi: 10.1099/mic.0.043257-0. Epub 2010 Aug 12. Microbiology (Reading). 2010. PMID: 20705664 Review.
-
Alterations of Gut Microbiota in Patients With Irritable Bowel Syndrome Based on 16S rRNA-Targeted Sequencing: A Systematic Review.Clin Transl Gastroenterol. 2019 Feb;10(2):e00012. doi: 10.14309/ctg.0000000000000012. Clin Transl Gastroenterol. 2019. PMID: 30829919 Free PMC article.
Cited by
-
Non-contiguous finished genome sequence and description of Alistipes timonensis sp. nov.Stand Genomic Sci. 2012 Jul 30;6(3):315-24. doi: 10.4056/sigs.2685971. Epub 2012 Jul 20. Stand Genomic Sci. 2012. PMID: 23408657 Free PMC article.
-
Maternal micronutrients can modify colonic mucosal microbiota maturation in murine offspring.Gut Microbes. 2012 Sep-Oct;3(5):426-33. doi: 10.4161/gmic.20697. Epub 2012 Jun 20. Gut Microbes. 2012. PMID: 22713270 Free PMC article.
-
Human Intestinal Barrier Function in Health and Disease.Clin Transl Gastroenterol. 2016 Oct 20;7(10):e196. doi: 10.1038/ctg.2016.54. Clin Transl Gastroenterol. 2016. PMID: 27763627 Free PMC article. Review.
-
Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome.Sci Rep. 2015 Aug 4;5:12693. doi: 10.1038/srep12693. Sci Rep. 2015. PMID: 26239401 Free PMC article.
-
An update on the use and investigation of probiotics in health and disease.Gut. 2013 May;62(5):787-96. doi: 10.1136/gutjnl-2012-302504. Epub 2013 Mar 8. Gut. 2013. PMID: 23474420 Free PMC article. Review.
References
-
- Apley J. The Child with Abdominal Pains. Blackwell Scientific. 1975
-
- Zuckerman B, Stevenson J, Bailey V. Stomachaches and headaches in a community sample of preschool children. Pediatrics. 1987;79:677–82. - PubMed
-
- Miele E, Simeone D, Marino A, et al. Functional gastrointestinal disorders in children: an Italian prospective survey. Pediatrics. 2004;114:73–8. - PubMed
-
- Thakkar K, Gilger MA, Shulman RJ, et al. EGD in children with abdominal pain: a systematic review. Am J Gastroenterol. 2007;102:654–61. - PubMed
-
- Saps M, Sztainberg M, Di Lorenzo C. A prospective community-based study of gastroenterological symptoms in school-age children. J Pediatr Gastroenterol Nutr. 2006;43:477–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

