Current perspectives on the phylogeny of Filoviridae
- PMID: 21742058
- PMCID: PMC7106080
- DOI: 10.1016/j.meegid.2011.06.017
Current perspectives on the phylogeny of Filoviridae
Abstract
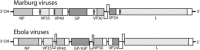
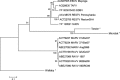
Sporadic fatal outbreaks of disease in humans and non-human primates caused by Ebola or Marburg viruses have driven research into the characterization of these viruses with the hopes of identifying host tropisms and potential reservoirs. Such an understanding of the relatedness of newly discovered filoviruses may help to predict risk factors for outbreaks of hemorrhagic disease in humans and/or non-human primates. Recent discoveries such as three distinct genotypes of Reston ebolavirus, unexpectedly discovered in domestic swine in the Philippines; as well as a new species, Bundibugyo ebolavirus; the recent discovery of Lloviu virus as a potential new genus, Cuevavirus, within Filoviridae; and germline integrations of filovirus-like sequences in some animal species bring new insights into the relatedness of filoviruses, their prevalence and potential for transmission to humans. These new findings reveal that filoviruses are more diverse and may have had a greater influence on the evolution of animals than previously thought. Herein we review these findings with regard to the implications for understanding the host range, prevalence and transmission of Filoviridae.
Published by Elsevier B.V.
Figures

References
-
- Anon, 2009. Imported case of Marburg hemorrhagic fever – Colorado, 2008. MMWR Morb Mortal Wkly Rep 58, 1377–1381. - PubMed
-
- Barrette R.W., Metwally S.A., Rowland J.M., Xu L., Zaki S.R., Nichol S.T., Rollin P.E., Towner J.S., Shieh W.J., Batten B., Sealy T.K., Carrillo C., Moran K.E., Bracht A.J., Mayr G.A., Sirios-Cruz M., Catbagan D.P., Lautner E.A., Ksiazek T.G., White W.R., McIntosh M.T. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325:204–206. - PubMed
-
- Dalgard D.W., Hardy R.J., Pearson S.L., Pucak G.J., Quander R.V., Zack P.M., Peters C.J., Jahrling P.B. Combined simian hemorrhagic fever and Ebola virus infection in cynomolgus monkeys. Lab. Anim. Sci. 1992;42:152–157. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

