Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1
- PMID: 21742260
- PMCID: PMC3134789
- DOI: 10.1016/j.str.2011.05.002
Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1
Abstract
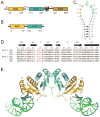
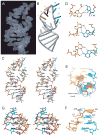
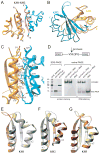
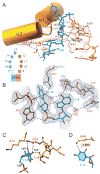
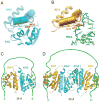
Nova onconeural antigens are neuron-specific RNA-binding proteins implicated in paraneoplastic opsoclonus-myoclonus-ataxia (POMA) syndrome. Nova harbors three K-homology (KH) motifs implicated in alternate splicing regulation of genes involved in inhibitory synaptic transmission. We report the crystal structure of the first two KH domains (KH1/2) of Nova-1 bound to an in vitro selected RNA hairpin, containing a UCAG-UCAC high-affinity binding site. Sequence-specific intermolecular contacts in the complex involve KH1 and the second UCAC repeat, with the RNA scaffold buttressed by interactions between repeats. Whereas the canonical RNA-binding surface of KH2 in the above complex engages in protein-protein interactions in the crystalline state, the individual KH2 domain can sequence-specifically target the UCAC RNA element in solution. The observed antiparallel alignment of KH1 and KH2 domains in the crystal structure of the complex generates a scaffold that could facilitate target pre-mRNA looping on Nova binding, thereby potentially explaining Nova's functional role in splicing regulation.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Albert ML, Darnell RB. Paraneoplastic neurological degenerations: keys to tumour immunity. Nat Rev Cancer. 2004;4:36–44. - PubMed
-
- Beuth B, Garcia-Mayoral MF, Taylor IA, Ramos A. Scaffold-independent analysis of RNA-protein interactions: the Nova-1 KH3-RNA complex. J Am Chem Soc. 2007;129:10205–10210. - PubMed
-
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- NS40955/NS/NINDS NIH HHS/United States
- GM07739/GM/NIGMS NIH HHS/United States
- HD40647/HD/NICHD NIH HHS/United States
- R01 NS034389/NS/NINDS NIH HHS/United States
- CA49982/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R56 NS034389/NS/NINDS NIH HHS/United States
- NS34389/NS/NINDS NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
- R01 CA049982/CA/NCI NIH HHS/United States
- R01 NS040955/NS/NINDS NIH HHS/United States
- R01 HD040647/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources

