Ultra-deep pyrosequencing analysis of the hepatitis B virus preCore region and main catalytic motif of the viral polymerase in the same viral genome
- PMID: 21742757
- PMCID: PMC3201856
- DOI: 10.1093/nar/gkr451
Ultra-deep pyrosequencing analysis of the hepatitis B virus preCore region and main catalytic motif of the viral polymerase in the same viral genome
Abstract
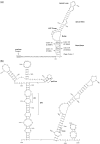
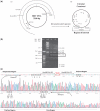
Hepatitis B virus (HBV) pregenomic RNA contains a hairpin structure (ε) located in the preCore region, essential for viral replication. ε stability is enhanced by the presence of preCore variants and ε is recognized by the HBV polymerase (Pol). Mutations in the retrotranscriptase domain (YMDD) of Pol are associated with treatment resistance. The aim of this study was to analyze the preCore region and YMDD motif by ultra-deep pyrosequencing (UDPS). To evaluate the UDPS error rate, an internal control sequence was inserted in the amplicon. A newly developed technique enabled simultaneous analysis of the preCore region and Pol in the same viral genome, as well as the conserved sequence of the internal control. Nucleotide errors in HindIII yielded a UDPS error rate <0.05%. UDPS study confirmed the possibility of simultaneous detection of preCore and YMDD mutations, and demonstrated the complexity of the HBV quasispecies and cooperation between viruses. Thermodynamic stability of the ε signal was found to be the main constraint for selecting main preCore mutations. Analysis of ε-signal variability suggested the essential nature of the ε structural motif and that certain nucleotides may be involved in ε signal functions.
Figures


Similar articles
-
Quasispecies dynamics in main core epitopes of hepatitis B virus by ultra-deep-pyrosequencing.World J Gastroenterol. 2012 Nov 14;18(42):6096-105. doi: 10.3748/wjg.v18.i42.6096. World J Gastroenterol. 2012. PMID: 23155338 Free PMC article.
-
Ultra-deep pyrosequencing detects conserved genomic sites and quantifies linkage of drug-resistant amino acid changes in the hepatitis B virus genome.PLoS One. 2012;7(5):e37874. doi: 10.1371/journal.pone.0037874. Epub 2012 May 30. PLoS One. 2012. PMID: 22666402 Free PMC article.
-
Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region.J Virol. 1993 Sep;67(9):5402-10. doi: 10.1128/JVI.67.9.5402-5410.1993. J Virol. 1993. PMID: 8350403 Free PMC article.
-
Quasispecies structure, cornerstone of hepatitis B virus infection: mass sequencing approach.World J Gastroenterol. 2013 Nov 7;19(41):6995-7023. doi: 10.3748/wjg.v19.i41.6995. World J Gastroenterol. 2013. PMID: 24222943 Free PMC article. Review.
-
Hepatitis B viruses: reverse transcription a different way.Virus Res. 2008 Jun;134(1-2):235-49. doi: 10.1016/j.virusres.2007.12.024. Epub 2008 Mar 12. Virus Res. 2008. PMID: 18339439 Review.
Cited by
-
Long-Term Follow-Up of Acute Hepatitis B: New Insights in Its Natural History and Implications for Antiviral Treatment.Genes (Basel). 2018 Jun 12;9(6):293. doi: 10.3390/genes9060293. Genes (Basel). 2018. PMID: 29895748 Free PMC article.
-
Extinction of hepatitis C virus by ribavirin in hepatoma cells involves lethal mutagenesis.PLoS One. 2013 Aug 16;8(8):e71039. doi: 10.1371/journal.pone.0071039. eCollection 2013. PLoS One. 2013. PMID: 23976977 Free PMC article.
-
Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data.Front Microbiol. 2012 Sep 11;3:329. doi: 10.3389/fmicb.2012.00329. eCollection 2012. Front Microbiol. 2012. PMID: 22973268 Free PMC article.
-
Initial sites of hepadnavirus integration into host genome in human hepatocytes and in the woodchuck model of hepatitis B-associated hepatocellular carcinoma.Oncogenesis. 2017 Apr 17;6(4):e317. doi: 10.1038/oncsis.2017.22. Oncogenesis. 2017. PMID: 28414318 Free PMC article.
-
Complex Genotype Mixtures Analyzed by Deep Sequencing in Two Different Regions of Hepatitis B Virus.PLoS One. 2015 Dec 29;10(12):e0144816. doi: 10.1371/journal.pone.0144816. eCollection 2015. PLoS One. 2015. PMID: 26714168 Free PMC article.
References
-
- Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235–249. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

