Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation
- PMID: 21742761
- PMCID: PMC3201876
- DOI: 10.1093/nar/gkr560
Structural basis for the biological relevance of the invariant apical stem in IRES-mediated translation
Abstract
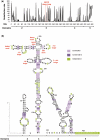
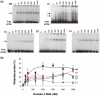
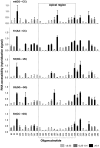
RNA structure plays a fundamental role in internal initiation of translation. Picornavirus internal ribosome entry site (IRES) are long, efficient cis-acting elements that recruit the ribosome to internal mRNA sites. However, little is known about long-range constraints determining the IRES RNA structure. Here, we sought to investigate the functional and structural relevance of the invariant apical stem of a picornavirus IRES. Mutation of this apical stem revealed better performance of G:C compared with C:G base pairs, demonstrating that the secondary structure solely is not sufficient for IRES function. In turn, mutations designed to disrupt the stem abolished IRES activity. Lack of tolerance to accept genetic variability in the apical stem was supported by the presence of coupled covariations within the adjacent stem-loops. SHAPE structural analysis, gel mobility-shift and microarrays-based RNA accessibility revealed that the apical stem contributes to maintain IRES RNA structure through the generation of distant interactions between two adjacent stem-loops. Our results demonstrate that a highly interactive structure constrained by distant interactions involving invariant G:C base pairs plays a key role in maintaining the RNA conformation necessary for IRES-mediated translation.
Figures



References
-
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol. Cell. 2010;40:228–237. - PubMed
-
- Vallejos M, Deforges J, Plank TD, Letelier A, Ramdohr P, Abraham CG, Valiente-Echeverria F, Kieft JS, Sargueil B, Lopez-Lastra M. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res. 2011 April 10; epub ahead of print; doi:10.1093/nar/gkr189. - PMC - PubMed

