Histone variant H3.3 stimulates HSP70 transcription through cooperation with HP1γ
- PMID: 21742762
- PMCID: PMC3201866
- DOI: 10.1093/nar/gkr529
Histone variant H3.3 stimulates HSP70 transcription through cooperation with HP1γ
Abstract
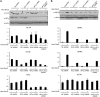
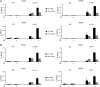
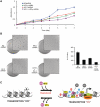
Histone variant H3.3 and heterochromatin protein 1γ (HP1γ) are two functional components of chromatin with role in gene transcription. However, the regulations of their dynamics during transcriptional activation and the molecular mechanisms underlying their actions remain poorly understood. Here, we provide evidence that heat shock-induced transcription of the human HSP70 gene is regulated via the coordinated and interdependent action of H3.3 and HP1γ. H3.3 and HP1γ are rapidly co-enriched at the human HSP70 promoters upon heat shock in a manner that closely parallels the initiation of transcription. Knockdown of H3.3 prevents the stable recruitment of HP1γ, inhibits active histone modifications, and attenuates HSP70 promoter activity. Likewise, knockdown of HP1γ leads to the decreased levels of H3.3 in the promoter regions and the repression of HSP70 genes. HP1γ selectively recognizes particular modification states of H3.3 in the nucleosome for its action. Moreover, HP1γ is overexpressed in three representative cancer cell lines, and its knockdown leads to reduction in HSP70 gene transcription and inhibition of cancer cell proliferation. We conclude that the physical and functional interactions between H3.3 and HP1γ make a unique contribution to acute HSP70 transcription and cancer development related to the misregulation of this transcription event.
Figures




References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 2008;9:15–26. - PubMed
-
- Albig W, Bramlage B, Gruber K, Klobeck HG, Kunz J, Doenecke D. The human replacement histone H3.3B gene (H3F3B) Genomics. 1995;30:264–272. - PubMed
-
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

