Staphylococcus aureus host range and human-bovine host shift
- PMID: 21742927
- PMCID: PMC3165375
- DOI: 10.1128/AEM.00238-11
Staphylococcus aureus host range and human-bovine host shift
Abstract
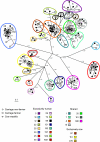
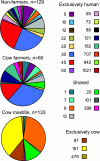
Staphylococcus aureus is a major agent of bovine mastitis. The concomitant emergence of pig-associated methicillin-resistant S. aureus (MRSA) in human carriage and infection requires a reexamination of the host range and specificity of human- and cow-associated S. aureus strains, something which has not been systematically studied previously. The genetic relatedness of 500 S. aureus isolates from bovine mastitis cases, 57 isolates from nasal carriage of farmers, and 133 isolates from nonfarmers was determined by amplified fragment length polymorphism (AFLP) analysis and spa typing. Multilocus sequence typing (MLST) was conducted on a subset of isolates to match AFLP clusters with MLST clonal complexes (CCs). This data set allowed us to study host range and host specificity and to estimate the extent of bovine-to-human transmission. The genotype compositions of S. aureus isolates from farmers and nonfarmers were very similar, while the mastitis isolates were quite distinct. Overall, transmission was low, but specific genotypes did show increased cow-to-human transmission. Unexpectedly, more than one-third of mastitis isolates belonged to CC8, a lineage which has not been considered to be bovine mastitis associated, but it is well known from human carriage and infection (i.e., USA300). Despite the fact that we did detect some transmission of other genotypes from cows to farmers, no transmission of CC8 isolates to farmers was detected, except for one tentative case. This was despite the close genetic relatedness of mastitis CC8 strains to nonfarmer carriage strains. These results suggest that the emergence of the new bovine-adapted genotype was due to a recent host shift from humans to cows concurrent with a loss of the ability to colonize humans. More broadly, our results indicate that host specificity is a lineage-specific trait that can rapidly evolve.
Figures


References
-
- Capurro A., Aspan A., Unnerstad H. E., Waller K. P., Artursson K. 2010. Identification of potential sources of Staphylococcus aureus in herds with mastitis problems. J. Dairy Sci. 93:180–191 - PubMed
-
- de Neeling A. J., et al. 2007. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 122:366–372 - PubMed
-
- Diep B. A., et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739 - PubMed
-
- Elphinstone M. S., Hinten G. N., Anderson M. J., Nock C. J. 2003. An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Mol. Ecol. Notes 3:317–320
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

