Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber
- PMID: 21742987
- PMCID: PMC3226218
- DOI: 10.1105/tpc.111.087189
Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber
Abstract
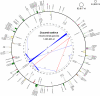
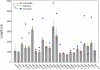
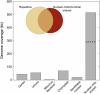
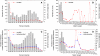
Members of the flowering plant family Cucurbitaceae harbor the largest known mitochondrial genomes. Here, we report the 1685-kb mitochondrial genome of cucumber (Cucumis sativus). We help solve a 30-year mystery about the origins of its large size by showing that it mainly reflects the proliferation of dispersed repeats, expansions of existing introns, and the acquisition of sequences from diverse sources, including the cucumber nuclear and chloroplast genomes, viruses, and bacteria. The cucumber genome has a novel structure for plant mitochondria, mapping as three entirely or largely autonomous circular chromosomes (lengths 1556, 84, and 45 kb) that vary in relative abundance over a twofold range. These properties suggest that the three chromosomes replicate independently of one another. The two smaller chromosomes are devoid of known functional genes but nonetheless contain diagnostic mitochondrial features. Paired-end sequencing conflicts reveal differences in recombination dynamics among chromosomes, for which an explanatory model is developed, as well as a large pool of low-frequency genome conformations, many of which may result from asymmetric recombination across intermediate-sized and sometimes highly divergent repeats. These findings highlight the promise of genome sequencing for elucidating the recombinational dynamics of plant mitochondrial genomes.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

