Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway
- PMID: 21742991
- PMCID: PMC3226213
- DOI: 10.1105/tpc.111.084608
Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway
Abstract
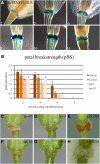
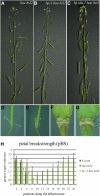
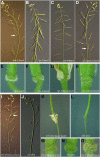
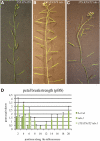
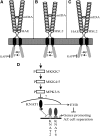
Floral organ abscission in Arabidopsis thaliana is regulated by the putative ligand-receptor system comprising the signaling peptide INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and the two receptor-like kinases HAESA and HAESA-LIKE2. The IDA signaling pathway presumably activates a MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) cascade to induce separation between abscission zone (AZ) cells. Misexpression of IDA effectuates precocious floral abscission and ectopic cell separation in latent AZ cell regions, which suggests that negative regulators are in place to prevent unrestricted and untimely AZ cell separation. Through a screen for mutations that restore floral organ abscission in ida mutants, we identified three new mutant alleles of the KNOTTED-LIKE HOMEOBOX gene BREVIPEDICELLUS (BP)/KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 (KNAT1). Here, we show that bp mutants, in addition to shedding their floral organs prematurely, have phenotypic commonalities with plants misexpressing IDA, such as enlarged AZ cells. We propose that BP/KNAT1 inhibits floral organ cell separation by restricting AZ cell size and number and put forward a model whereby IDA signaling suppresses BP/KNAT1, which in turn allows KNAT2 and KNAT6 to induce floral organ abscission.
Figures


References
-
- Aalen R.B. (2011). Flower and floral organ abscission: Control, gene expression and hormone interaction. In The Flowering Process and Its Control in Plants: Gene Expression and Hormone Interaction, Yaish M.W., (Kerala, India: Research Signpost/Transworld Research Network; ), pp. 307–327
-
- Alonso-Cantabrana H., Ripoll J.J., Ochando I., Vera A., Ferrándiz C., Martínez-Laborda A. (2007). Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 134: 2663–2671 - PubMed
-
- Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561: 127–131 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

