CYP3A5, ABCB1, and SLCO1B1 polymorphisms and pharmacokinetics and virologic outcome of lopinavir/ritonavir in HIV-infected children
- PMID: 21743379
- PMCID: PMC3139013
- DOI: 10.1097/FTD.0b013e318225384f
CYP3A5, ABCB1, and SLCO1B1 polymorphisms and pharmacokinetics and virologic outcome of lopinavir/ritonavir in HIV-infected children
Abstract
Objective: CYP3A5, MDR1 (ABCB1), and OATP1 (SLCO1B1) polymorphisms have been associated with variability in the pharmacokinetics (PK) of protease inhibitors. The aim of this study was to investigate the influence of CYP3A5 A6986G, ABCB1 (C3435T and G2677T), and SLCO1B1 (T521C and A388AG) polymorphisms on the PK and virologic outcome of lopinavir/ritonavir (LPV/RTV) in HIV-infected children.
Design and methods: We conducted a prospective cohort study in children (4-18 years old) on stable antiretroviral therapy with LPV/RTV. CYP3A5, ABCB1, and SLCO1B1 genotypes were determined using polymerase chain reaction amplification with allelic discrimination assays. The 12-hour plasma area under the concentration-time curves (AUC) and clearances (CL) of LPV and RTV were estimated using noncompartmental models. HIV RNA viral load was evaluated every 12 weeks for a total study period of 52 weeks. Analysis of covariance models with adjustment for age and adherence and allometric adjustment of CL were used to assess associations between studied polymorphisms and AUC, CL, and HIV RNA.
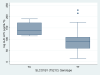
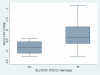
Results: Fifty children (median age 11.2 years) were enrolled. Allele frequencies of the genotypes studied were in Hardy-Weinberg equilibrium. There was no statistically significant association between LPV or RTV AUC or CL, and CYP3A5, ABCB1, or SLCO1B1 A388G polymorphisms. There was a significant association between SLCO1B1 T521C genotype and increased LPV AUC (P = 0.042) and a nearly significant association with decreased LPV CL (P = 0.063). None of the studied polymorphisms, including SLCO1B1 T521C, were associated with virologic outcome during 52 weeks of study follow-up.
Conclusions: There was no statistically significant influence of the CYP3A5, ABCB1, or SLCO1B1 A388AG polymorphisms on the PK and virologic outcome of LPV/RTV in HIV-infected children. SLCO1B1 T521C polymorphism was significantly associated with an increase in LPV AUC but was not associated with undetectable HIV RNA during the study period.
Figures


References
-
- Mouly SJ, Matheny C, Paine MF, Smith G, Lamba J, Lamba V, et al. Variation in oral clearance of saquinavir is predicted by CYP3A5*1 genotype but not by enterocyte content of cytochrome P450 3A5. Clin Pharmacol Ther. 2005;78:605–618. - PubMed
-
- Weemhoff JL, von Moltke LL, Richert C, Hesse LM, Harmatz JS, Greenblatt DJ. Apparent mechanism-based inhibition of human CYP3A in-vitro by lopinavir. J Pharm Pharmacol. 2003;55:381–386. - PubMed
-
- Josephson F, Allqvist A, Janabi M, Sayi J, Aklillu E, Jande M, et al. CYP3A5 genotype has an impact on the metabolism of the HIV protease inhibitor saquinavir. Clin Pharmacol Ther. 2007;81:708–712. - PubMed
-
- Woodahl EL, Yang Z, Bui T, Shen DD, Ho RJ. MDR1 G1199A polymorphism alters permeability of HIV protease inhibitors across P-glycoprotein-expressing epithelial cells. Aids. 2005;19:1617–1625. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K23 AI076106-01/AI/NIAID NIH HHS/United States
- K23 AI076106/AI/NIAID NIH HHS/United States
- K12 RR017613/RR/NCRR NIH HHS/United States
- U10 HD045993/HD/NICHD NIH HHS/United States
- 1K12 RR017613/RR/NCRR NIH HHS/United States
- 5K24RR019729/RR/NCRR NIH HHS/United States
- 1U10 HD45993/HD/NICHD NIH HHS/United States
- K23 HD060452/HD/NICHD NIH HHS/United States
- K24 RR019729/RR/NCRR NIH HHS/United States
- M01 RR020359/RR/NCRR NIH HHS/United States
- 5 K23 HD060452-02/HD/NICHD NIH HHS/United States
- M01-RR-020359/RR/NCRR NIH HHS/United States
- R01 EB005803/EB/NIBIB NIH HHS/United States
- R01 EB005803-01A1/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

