Increased gene copy number of ERG on chromosome 21 but not TMPRSS2-ERG fusion predicts outcome in prostatic adenocarcinomas
- PMID: 21743434
- PMCID: PMC3360950
- DOI: 10.1038/modpathol.2011.111
Increased gene copy number of ERG on chromosome 21 but not TMPRSS2-ERG fusion predicts outcome in prostatic adenocarcinomas
Abstract
The role of TMPRSS2-ERG gene fusion in prostate cancer prognostication remains controversial. We evaluated the prognostic role of TMPRSS2-ERG fusion using fluorescence in situ hybridization analysis in a case-control study nested in The Johns Hopkins retropubic radical prostatectomy cohort. In all, 10 tissue microarrays containing paired tumors and normal tissues obtained from 172 cases (recurrence) and 172 controls (non-recurrence) matched on pathological grade, stage, race/ethnicity, and age at the time of surgery were analyzed. All radical prostatectomies were performed at our institution between 1993 and 2004. Recurrence was defined as biochemical recurrence, development of clinical evidence of metastasis, or death from prostate carcinoma. Each tissue microarray spot was scored for the presence of TMPRSS2-ERG gene fusion and for ERG gene copy number gains. The odds ratio of recurrence and 95% confidence intervals were estimated from conditional logistic regression. Although the percentage of cases with fusion was slightly lower in cases than in controls (50 vs 57%), the difference was not statistically significant (P=0.20). The presence of fusion due to either deletion or split event was not associated with recurrence. Similarly, the presence of duplicated ERG deletion, duplicated ERG split, or ERG gene copy number gain with a single ERG fusion was not associated with recurrence. ERG gene polysomy without fusion was significantly associated with recurrence (odds ratio 2.0, 95% confidence interval 1.17-3.42). In summary, TMPRSS2-ERG fusion was not prognostic for recurrence after retropubic radical prostatectomy for clinically localized prostate cancer, although men with ERG gene copy number gain without fusion were twice more likely to recur.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

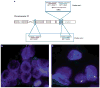

References
-
- Schroder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. - PubMed
-
- Perner S, Mosquera JM, Demichelis F, et al. TMPRSS2- ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

