PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution
- PMID: 21743438
- PMCID: PMC3160651
- DOI: 10.1038/emboj.2011.226
PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution
Abstract
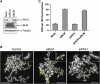
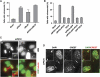
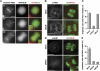
Centromeres nucleate the formation of kinetochores and are vital for chromosome segregation during mitosis. The SNF2 family helicase PICH (Plk1-interacting checkpoint helicase) and the BLM (the Bloom's syndrome protein) helicase decorate ultrafine histone-negative DNA threads that link the segregating sister centromeres during anaphase. The functions of PICH and BLM at these threads are not understood, however. Here, we show that PICH binds to BLM and enables BLM localization to anaphase centromeric threads. PICH- or BLM-RNAi cells fail to resolve these threads in anaphase. The fragmented threads form centromeric-chromatin-containing micronuclei in daughter cells. Anaphase threads in PICH- and BLM-RNAi cells contain histones and centromere markers. Recombinant purified PICH has nucleosome remodelling activities in vitro. We propose that PICH and BLM unravel centromeric chromatin and keep anaphase DNA threads mostly free of nucleosomes, thus allowing these threads to span long distances between rapidly segregating centromeres without breakage and providing a spatiotemporal window for their resolution.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

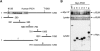


References
-
- Baumann C, Korner R, Hofmann K, Nigg EA (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128: 101–114 - PubMed
-
- Bharadwaj R, Yu H (2004) The spindle checkpoint, aneuploidy, and cancer. Oncogene 23: 2016–2027 - PubMed
-
- Buscaino A, Allshire R, Pidoux A (2010) Building centromeres: home sweet home or a nomadic existence? Curr Opin Genet Dev 20: 118–126 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

