Mir193b-365 is essential for brown fat differentiation
- PMID: 21743466
- PMCID: PMC3149720
- DOI: 10.1038/ncb2286
Mir193b-365 is essential for brown fat differentiation
Abstract
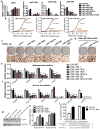
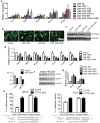
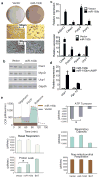
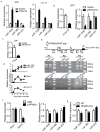
Mammals have two principal types of fat. White adipose tissue primarily serves to store extra energy as triglycerides, whereas brown adipose tissue is specialized to burn lipids for heat generation and energy expenditure as a defence against cold and obesity. Recent studies have demonstrated that brown adipocytes arise in vivo from a Myf5-positive, myoblastic progenitor by the action of Prdm16 (PR domain containing 16). Here, we identified a brown-fat-enriched miRNA cluster, MiR-193b-365, as a key regulator of brown fat development. Blocking miR-193b and/or miR-365 in primary brown preadipocytes markedly impaired brown adipocyte adipogenesis by enhancing Runx1t1 (runt-related transcription factor 1; translocated to, 1) expression, whereas myogenic markers were significantly induced. Forced expression of Mir193b and/or Mir365 in C2C12 myoblasts blocked the entire programme of myogenesis, and, in adipogenic conditions, miR-193b induced myoblasts to differentiate into brown adipocytes. Mir193b-365 was upregulated by Prdm16 partially through Pparα. Our results demonstrate that Mir193b-365 serves as an essential regulator for brown fat differentiation, in part by repressing myogenesis.
Conflict of interest statement
The authors declare no financial conflict of interest.
Figures

References
-
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. - PubMed
-
- van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. - PubMed
-
- Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

