Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate
- PMID: 21743471
- PMCID: PMC3144989
- DOI: 10.1038/nn.2859
Neurod6 expression defines new retinal amacrine cell subtypes and regulates their fate
Abstract
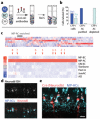
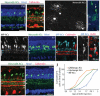
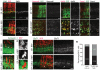
Most regions of the CNS contain many subtypes of inhibitory interneurons with specialized roles in circuit function. In the mammalian retina, the ∼30 subtypes of inhibitory interneurons called amacrine cells (ACs) are generally divided into two groups: wide/medium-field GABAergic ACs and narrow-field glycinergic ACs, which mediate lateral and vertical interactions, respectively, within the inner plexiform layer. We used expression profiling and mouse transgenic lines to identify and characterize two closely related narrow-field AC subtypes. Both arise postnatally and one is neither glycinergic nor GABAergic (nGnG). Two transcription factors selectively expressed by these subtypes, Neurod6 and special AT-rich-sequence-binding protein 2 (Satb2), regulate a postmitotic cell fate choice between these subtypes. Satb2 induces Neurod6, which persists in nGnG ACs and promotes their fate but is downregulated in the related glycinergic AC subtype. Our results support the view that cell fate decisions made in progenitors and their progeny act together to diversify ACs.
Figures




References
-
- Cossart R. The maturation of cortical interneuron diversity: how multiple developmental journeys shape the emergence of proper network function. Curr Opin Neurobiol. epub 2010. - PubMed
-
- Goulding M, Pfaff SL. Development of circuits that generate simple rhythmic behaviors in vertebrates. Curr Opin Neurobiol. 2005;15:14–20. - PubMed
-
- MacNeil MA, Masland RH. Extreme diversity among amacrine cells: implications for function. Neuron. 1998;20:971–982. - PubMed
-
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol. 2004;480:331–351. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

