Genetics of sweet taste preferences
- PMID: 21743773
- PMCID: PMC3130742
- DOI: 10.1002/ffj.2074
Genetics of sweet taste preferences
Abstract
Sweet taste is a powerful factor influencing food acceptance. There is considerable variation in sweet taste perception and preferences within and among species. Although learning and homeostatic mechanisms contribute to this variation in sweet taste, much of it is genetically determined. Recent studies have shown that variation in the T1R genes contributes to within- and between-species differences in sweet taste. In addition, our ongoing studies using the mouse model demonstrate that a significant portion of variation in sweetener preferences depends on genes that are not involved in peripheral taste processing. These genes are likely involved in central mechanisms of sweet taste processing, reward and/or motivation. Genetic variation in sweet taste not only influences food choice and intake, but is also associated with proclivity to drink alcohol. Both peripheral and central mechanisms of sweet taste underlie correlation between sweet-liking and alcohol consumption in animal models and humans. All these data illustrate complex genetics of sweet taste preferences and its impact on human nutrition and health. Identification of genes responsible for within- and between-species variation in sweet taste can provide tools to better control food acceptance in humans and other animals.
Figures

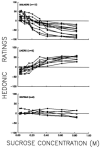
References
-
- Teff KL, Devine J, Engelman K. Physiol. Behav. 1995;57:1089. - PubMed
-
- Grill HJ, Berridge KC, Ganster DJ. Am. J. Physiol. 1984;246:R88. - PubMed
-
- Malaisse WJ, Vanonderbergen A, Louchami K, Jijakli H, Malaisse-Lagae F. Cell. Signal. 1998;10:727. - PubMed
-
- Yamamoto T, Sako N, Maeda S. Physiol. Behav. 2000;69:345. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
