Plants versus pathogens: an evolutionary arms race
- PMID: 21743794
- PMCID: PMC3131095
- DOI: 10.1071/FP09304
Plants versus pathogens: an evolutionary arms race
Abstract
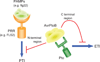
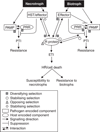
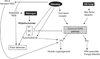
The analysis of plant-pathogen interactions is a rapidly moving research field and one that is very important for productive agricultural systems. The focus of this review is on the evolution of plant defence responses and the coevolution of their pathogens, primarily from a molecular-genetic perspective. It explores the evolution of the major types of plant defence responses including pathogen associated molecular patterns and effector triggered immunity as well as the forces driving pathogen evolution, such as the mechanisms by which pathogen lineages and species evolve. Advances in our understanding of plant defence signalling, stomatal regulation, R gene-effector interactions and host specific toxins are used to highlight recent insights into the coevolutionary arms race between pathogens and plants. Finally, the review considers the intriguing question of how plants have evolved the ability to distinguish friends such as rhizobia and mycorrhiza from their many foes.
Figures
References
-
- Altenbach D, Robatzek S. Pattern recognition receptors: from the cell surface to intracellular dynamics. Molecular Plant–Microbe Interactions. 2007;20:1031–1039. doi:10.1094/MPMI-20-9-1031. - PubMed
-
- Araki H, Tian D, Goss EM, Jakob K, Halldorsdottir SS, Kreitman M, Bergelson J. Presence/absence polymorphism for alternative pathogenicity islands in Pseudomonas viridiflava, a pathogen of Arabidopsis; Proceedings of the National Academy of Sciences of the United States of America; 2006. pp. 5887–5892. doi:10.1073/pnas.0601431103. - PMC - PubMed
-
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi:10.1038/415977a. - PubMed
-
- Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi:10.1016/S0092-8674(03)00036-9. - PubMed
-
- Bari R, Jones J. Role of plant hormones in plant defence responses. Plant Molecular Biology. 2009;69:473–488. doi:10.1007/s11103-008-9435-0. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

