Hes-1 regulates the excitatory fate of neural progenitors through modulation of Tlx3 (HOX11L2) expression
- PMID: 21744064
- PMCID: PMC11114997
- DOI: 10.1007/s00018-011-0765-8
Hes-1 regulates the excitatory fate of neural progenitors through modulation of Tlx3 (HOX11L2) expression
Abstract
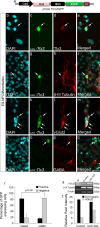
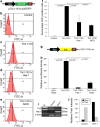
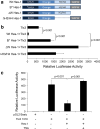
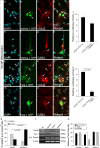
Tlx3 (HOX11L2) is regarded as one of the selector genes in excitatory versus inhibitory fate specification of neurons in distinct regions of the nervous system. Expression of Tlx3 in a post-mitotic immature neuron favors a glutamatergic over GABAergic fate. The factors that regulate Tlx3 have immense importance in the fate specification of glutamatergic neurons. Here, we have shown that Notch target gene, Hes-1, negatively regulates Tlx3 expression, resulting in decreased generation of glutamatergic neurons. Down-regulation of Hes-1 removed the inhibition on Tlx3 promoter, thus promoting glutamatergic differentiation. Promoter-protein interaction studies with truncated/mutated Hes-1 protein suggested that the co-repressor recruitment mediated through WRPW domain of Hes-1 has contributed to the repressive effect. Our results clearly demonstrate a new and unique role for canonical Notch signaling through Hes-1, in neurotransmitter/subtype fate specification of neurons in addition to its known functional role in proliferation/maintenance of neural progenitors.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

