A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology
- PMID: 21744844
- PMCID: PMC3522851
- DOI: 10.1021/bi200805j
A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology
Abstract
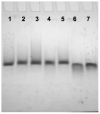
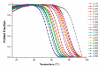
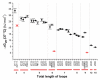
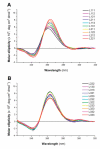
G-Quadruplexes are noncanonical nucleic acid secondary structures based on guanine association that are readily adopted by G-rich RNA and DNA sequences. Naturally occurring genomic G-quadruplex-forming sequences have functional roles in biology that are mediated through structure. To appreciate how this is achieved, an understanding of the likelihood of G-quadruplex formation and the structural features of the folded species under a defined set of conditions is informative. We previously systematically investigated the thermodynamic stability and folding topology of DNA G-quadruplexes and found a strong dependence of these properties on loop length and loop arrangement [Bugaut, A., and Balasubramanian, S. (2008) Biochemistry 47, 689-697]. Here we report on a complementary analysis of RNA G-quadruplexes using UV melting and circular dichroism spectroscopy that also serves as a comparison to the equivalent DNA G-quadruplex-forming sequences. We found that the thermodynamic stability of G-quadruplex RNA can be modulated by loop length while the overall structure is largely unaffected. The systematic design of our study also revealed subtle loop length dependencies in RNA G-quadruplex structure.
© 2011 American Chemical Society
Figures
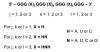
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

