Reconstitution of a functional IS608 single-strand transpososome: role of non-canonical base pairing
- PMID: 21745812
- PMCID: PMC3201880
- DOI: 10.1093/nar/gkr566
Reconstitution of a functional IS608 single-strand transpososome: role of non-canonical base pairing
Abstract
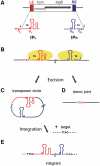
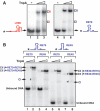
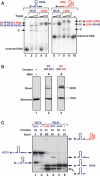
Single-stranded (ss) transposition, a recently identified mechanism adopted by members of the widespread IS200/IS605 family of insertion sequences (IS), is catalysed by the transposase, TnpA. The transposase of IS608, recognizes subterminal imperfect palindromes (IP) at both IS ends and cleaves at sites located at some distance. The cleavage sites, C, are not recognized directly by the protein but by short sequences 5' to the foot of each IP, guide (G) sequences, using a network of canonical ('Watson-Crick') base interactions. In addition a set of non-canonical base interactions similar to those found in RNA structures are also involved. We have reconstituted a biologically relevant complex, the transpososome, including both left and right ends and TnpA, which catalyses excision of a ss DNA circle intermediate. We provide a detailed picture of the way in which the IS608 transpososome is assembled and demonstrate that both C and G sequences are essential for forming a robust transpososome detectable by EMSA. We also address several questions central to the organization and function of the ss transpososome and demonstrate the essential role of non-canonical base interactions in the IS608 ends for its stability by using point mutations which destroy individual non-canonical base interactions.
Figures


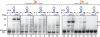
Similar articles
-
In vitro reconstitution of a single-stranded transposition mechanism of IS608.Mol Cell. 2008 Feb 15;29(3):302-12. doi: 10.1016/j.molcel.2007.12.008. Mol Cell. 2008. PMID: 18280236
-
IS200/IS605 family single-strand transposition: mechanism of IS608 strand transfer.Nucleic Acids Res. 2013 Mar 1;41(5):3302-13. doi: 10.1093/nar/gkt014. Epub 2013 Jan 23. Nucleic Acids Res. 2013. PMID: 23345619 Free PMC article.
-
Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection.Cell. 2008 Jan 25;132(2):208-20. doi: 10.1016/j.cell.2007.12.029. Cell. 2008. PMID: 18243097 Free PMC article.
-
The IS200/IS605 Family and "Peel and Paste" Single-strand Transposition Mechanism.Microbiol Spectr. 2015 Aug;3(4). doi: 10.1128/microbiolspec.MDNA3-0039-2014. Microbiol Spectr. 2015. PMID: 26350330 Review.
-
Mariner and the ITm Superfamily of Transposons.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0033-2014.. doi: 10.1128/microbiolspec.MDNA3-0033-2014. Microbiol Spectr. 2015. PMID: 26104691 Review.
Cited by
-
Flexible TAM requirement of TnpB enables efficient single-nucleotide editing with expanded targeting scope.Nat Commun. 2024 Apr 24;15(1):3464. doi: 10.1038/s41467-024-47697-4. Nat Commun. 2024. PMID: 38658536 Free PMC article.
-
Single-strand DNA processing: phylogenomics and sequence diversity of a superfamily of potential prokaryotic HuH endonucleases.BMC Genomics. 2018 Jun 19;19(1):475. doi: 10.1186/s12864-018-4836-1. BMC Genomics. 2018. PMID: 29914351 Free PMC article.
-
Targeting IS608 transposon integration to highly specific sequences by structure-based transposon engineering.Nucleic Acids Res. 2018 May 4;46(8):4152-4163. doi: 10.1093/nar/gky235. Nucleic Acids Res. 2018. PMID: 29635476 Free PMC article.
-
Survey of chimeric IStron elements in bacterial genomes: multiple molecular symbioses between group I intron ribozymes and DNA transposons.Nucleic Acids Res. 2014 Nov 10;42(20):12333-51. doi: 10.1093/nar/gku939. Epub 2014 Oct 16. Nucleic Acids Res. 2014. PMID: 25324310 Free PMC article.
-
3dRNAscore: a distance and torsion angle dependent evaluation function of 3D RNA structures.Nucleic Acids Res. 2015 May 26;43(10):e63. doi: 10.1093/nar/gkv141. Epub 2015 Feb 24. Nucleic Acids Res. 2015. PMID: 25712091 Free PMC article.
References
-
- Siguier P, Filee J, Chandler M. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 2006;9:526–531. - PubMed
-
- Guynet C, Hickman AB, Barabas O, Dyda F, Chandler M, Ton-Hoang B. In vitro reconstitution of a single-stranded transposition mechanism of IS608. Mol. Cell. 2008;29:302–312. - PubMed
-
- Ronning DR, Guynet C, Ton-Hoang B, Perez ZN, Ghirlando R, Chandler M, Dyda F. Active site sharing and subterminal hairpin recognition in a new class of DNA transposases. Mol. Cell. 2005;20:143–154. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

