Development of full-length cDNAs from Chinese cabbage (Brassica rapa Subsp. pekinensis) and identification of marker genes for defence response
- PMID: 21745830
- PMCID: PMC3158467
- DOI: 10.1093/dnares/dsr018
Development of full-length cDNAs from Chinese cabbage (Brassica rapa Subsp. pekinensis) and identification of marker genes for defence response
Abstract
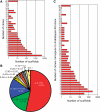
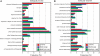
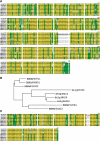
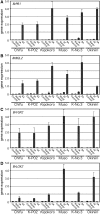
Arabidopsis belongs to the Brassicaceae family and plays an important role as a model plant for which researchers have developed fine-tuned genome resources. Genome sequencing projects have been initiated for other members of the Brassicaceae family. Among these projects, research on Chinese cabbage (Brassica rapa subsp. pekinensis) started early because of strong interest in this species. Here, we report the development of a library of Chinese cabbage full-length cDNA clones, the RIKEN BRC B. rapa full-length cDNA (BBRAF) resource, to accelerate research on Brassica species. We sequenced 10 000 BBRAF clones and confirmed 5476 independent clones. Most of these cDNAs showed high homology to Arabidopsis genes, but we also obtained more than 200 cDNA clones that lacked any sequence homology to Arabidopsis genes. We also successfully identified several possible candidate marker genes for plant defence responses from our analysis of the expression of the Brassica counterparts of Arabidopsis marker genes in response to salicylic acid and jasmonic acid. We compared gene expression of these markers in several Chinese cabbage cultivars. Our BBRAF cDNA resource will be publicly available from the RIKEN Bioresource Center and will help researchers to transfer Arabidopsis-related knowledge to Brassica crops.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

