A pathway for the control of anoikis sensitivity by E-cadherin and epithelial-to-mesenchymal transition
- PMID: 21746881
- PMCID: PMC3187352
- DOI: 10.1128/MCB.01342-10
A pathway for the control of anoikis sensitivity by E-cadherin and epithelial-to-mesenchymal transition
Abstract
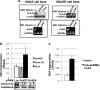
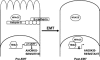
Detachment of epithelial cells from matrix or attachment to an inappropriate matrix engages an apoptotic response known as anoikis, which prevents metastasis. Cellular sensitivity to anoikis is compromised during the oncogenic epithelial-to-mesenchymal transition (EMT), through unknown mechanisms. We report here a pathway through which EMT confers anoikis resistance. NRAGE (neurotrophin receptor-interacting melanoma antigen) interacted with a component of the E-cadherin complex, ankyrin-G, maintaining NRAGE in the cytoplasm. Oncogenic EMT downregulated ankyrin-G, enhancing the nuclear localization of NRAGE. The oncogenic transcriptional repressor protein TBX2 interacted with NRAGE, repressing the tumor suppressor gene p14ARF. P14ARF sensitized cells to anoikis; conversely, the TBX2/NRAGE complex protected cells against anoikis by downregulating this gene. This represents a novel pathway for the regulation of anoikis by EMT and E-cadherin.
Figures








References
-
- Ayyanathan K., et al. 2007. The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic shuttling. Cancer Res. 67:9097–9106 - PubMed
-
- Barker P. A., Salehi A. 2002. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci. Res. 67:705–712 - PubMed
-
- Barrallo-Gimeno A., Nieto M. A. 2005. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132:3151–3161 - PubMed
-
- Bartel P. L., Fields S. 1995. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254:241–263 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
