Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids
- PMID: 21746892
- PMCID: PMC3158150
- DOI: 10.1073/pnas.1103190108
Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids
Abstract
Uniparental chromosome elimination occurs in several interspecific hybrids of plants. We studied the mechanism underlying selective elimination of the paternal chromosomes during the early development of Hordeum vulgare × Hordeum bulbosum embryos. The following conclusions regarding the role of the centromere-specific histone H3 variant (CENH3) in the process of chromosome elimination were drawn: (i) centromere inactivity of H. bulbosum chromosomes triggers the mitosis-dependent process of uniparental chromosome elimination in unstable H. vulgare × H. bulbosum hybrids; (ii) centromeric loss of CENH3 protein rather than uniparental silencing of CENH3 genes causes centromere inactivity; (iii) in stable species combinations, cross-species incorporation of CENH3 occurs despite centromere-sequence differences, and not all CENH3 variants get incorporated into centromeres if multiple CENH3s are present in species combinations; and (iv) diploid barley species encode two CENH3 variants, the proteins of which are intermingled within centromeres throughout mitosis and meiosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



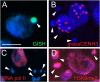



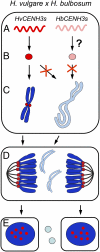
Comment in
-
In a battle between parental chromosomes, a failure to reload.Proc Natl Acad Sci U S A. 2011 Aug 16;108(33):13361-2. doi: 10.1073/pnas.1110320108. Epub 2011 Aug 3. Proc Natl Acad Sci U S A. 2011. PMID: 21813763 Free PMC article. No abstract available.
References
-
- Kasha KJ, Kao KN. High frequency haploid production in barley (Hordeum vulgare L.) Nature. 1970;225:874–876. - PubMed
-
- Devaux P, Pickering R. Haploids in the improvement of poaceae. In: Palmer CE, Keller WA, Kasha KJ, editors. Haploids in Crop Improvement II, Biotechnology in Agriculture and Forestry Series. Vol. 56. Berlin: Springer; 2005. pp. 215–242.
-
- Bennett MD, Finch RA, Barclay IR. The time rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma. 1976;54:175–200.
-
- Gernand D, Rutten T, Pickering R, Houben A. Elimination of chromosomes in Hordeum vulgare x H. bulbosum crosses at mitosis and interphase involves micronucleus formation and progressive heterochromatinization. Cytogenet Genome Res. 2006;114:169–174. - PubMed
-
- Subrahmanyam NC, Kasha KJ. Selective chromosome elimination during haploid formation in barley following interspecific hybridization. Chromosoma. 1973;42:111–112.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

