Novel postentry inhibitor of human immunodeficiency virus type 1 replication screened by yeast membrane-associated two-hybrid system
- PMID: 21746942
- PMCID: PMC3165301
- DOI: 10.1128/AAC.00299-11
Novel postentry inhibitor of human immunodeficiency virus type 1 replication screened by yeast membrane-associated two-hybrid system
Abstract
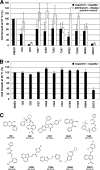
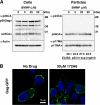
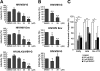
Human immunodeficiency virus (HIV) Gag protein targets to the plasma membrane and assembles into viral particles. In the next round of infection, the mature Gag capsids disassemble during viral entry. Thus, Gag plays a central role in the HIV life cycle. Using a yeast membrane-associated two-hybrid assay based on the SOS-RAS signaling system, we developed a system to measure the Gag-Gag interaction and isolated 6 candidates for Gag assembly inhibitors from a chemical library composed of 20,000 small molecules. When tested in the human MT-4 cell line and primary peripheral blood mononuclear cells, one of the candidates, 2-(benzothiazol-2-ylmethylthio)-4-methylpyrimidine (BMMP), displayed an inhibitory effect on HIV replication, although a considerably high dose was required. Unexpectedly, neither particle production nor maturation was inhibited by BMMP. Confocal microscopy confirmed that BMMP did not block Gag plasma membrane targeting. Single-round infection assays with envelope-pseudotyped and luciferase-expressing viruses revealed that BMMP inhibited HIV replication postentry but not simian immunodeficiency virus (SIV) or murine leukemia virus infection. Studies with HIV/SIV Gag chimeras indicated that the Gag capsid (CA) domain was responsible for the BMMP-mediated HIV postentry block. In vitro studies indicated that BMMP accelerated disassembly of HIV cores and, conversely, inhibited assembly of purified CA protein in a dose-dependent manner. Collectively, our data suggest that BMMP primarily targets the HIV CA domain and disrupts viral infection postentry, possibly through inducing premature disassembly of HIV cores. We suggest that BMMP is a potential lead compound to develop antiretroviral drugs bearing novel mechanisms of action.
Figures




References
-
- Aronheim A., et al. 1994. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell 78:949–961 - PubMed
-
- Bartonova V., et al. 2008. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J. Biol. Chem. 283:32024–32033 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

