Increases in p53 expression induce CTGF synthesis by mouse and human hepatocytes and result in liver fibrosis in mice
- PMID: 21747166
- PMCID: PMC3148730
- DOI: 10.1172/JCI44957
Increases in p53 expression induce CTGF synthesis by mouse and human hepatocytes and result in liver fibrosis in mice
Abstract
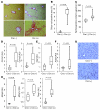
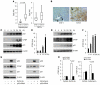
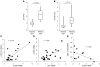
The tumor suppressor p53 has been implicated in the pathogenesis of non-cancer-related conditions such as insulin resistance, cardiac failure, and early aging. In addition, accumulation of p53 has been observed in the hepatocytes of individuals with fibrotic liver diseases, but the significance of this is not known. Herein, we have mechanistically linked p53 activation in hepatocytes to liver fibrosis. Hepatocyte-specific deletion in mice of the gene encoding Mdm2, a protein that promotes p53 degradation, led to hepatocyte synthesis of connective tissue growth factor (CTGF; the hepatic fibrogenic master switch), increased hepatocyte apoptosis, and spontaneous liver fibrosis; concurrent removal of p53 completely abolished this phenotype. Compared with wild-type controls, mice with hepatocyte-specific p53 deletion exhibited similar levels of hepatocyte apoptosis but decreased liver fibrosis and hepatic CTGF expression in two models of liver fibrosis. The clinical significance of these data was highlighted by two observations. First, p53 upregulated CTGF in a human hepatocellular carcinoma cell line by repressing miR-17-92. Second, human liver samples showed a correlation between CTGF and p53-regulated gene expression, which were both increased in fibrotic livers. This study reveals that p53 induces CTGF expression and promotes liver fibrosis, suggesting that the p53/CTGF pathway may be a therapeutic target in the treatment of liver fibrosis.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

