Epigenome-wide association studies for common human diseases
- PMID: 21747404
- PMCID: PMC3508712
- DOI: 10.1038/nrg3000
Epigenome-wide association studies for common human diseases
Abstract
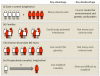
Despite the success of genome-wide association studies (GWASs) in identifying loci associated with common diseases, a substantial proportion of the causality remains unexplained. Recent advances in genomic technologies have placed us in a position to initiate large-scale studies of human disease-associated epigenetic variation, specifically variation in DNA methylation. Such epigenome-wide association studies (EWASs) present novel opportunities but also create new challenges that are not encountered in GWASs. We discuss EWAS design, cohort and sample selections, statistical significance and power, confounding factors and follow-up studies. We also discuss how integration of EWASs with GWASs can help to dissect complex GWAS haplotypes for functional analysis.
Figures

References
-
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–7. - PubMed
-
- Kulis M, Esteller M. DNA methylation and cancer. Advances in genetics. 2010;70:27–56. - PubMed
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:66981. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

