Systematic approaches towards the development of host-directed antiviral therapeutics
- PMID: 21747723
- PMCID: PMC3131607
- DOI: 10.3390/ijms12064027
Systematic approaches towards the development of host-directed antiviral therapeutics
Abstract
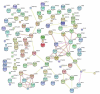
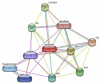
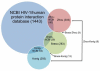
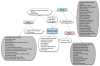
Since the onset of antiviral therapy, viral resistance has compromised the clinical value of small-molecule drugs targeting pathogen components. As intracellular parasites, viruses complete their life cycle by hijacking a multitude of host-factors. Aiming at the latter rather than the pathogen directly, host-directed antiviral therapy has emerged as a concept to counteract evolution of viral resistance and develop broad-spectrum drug classes. This approach is propelled by bioinformatics analysis of genome-wide screens that greatly enhance insights into the complex network of host-pathogen interactions and generate a shortlist of potential gene targets from a multitude of candidates, thus setting the stage for a new era of rational identification of drug targets for host-directed antiviral therapies. With particular emphasis on human immunodeficiency virus and influenza virus, two major human pathogens, we review screens employed to elucidate host-pathogen interactions and discuss the state of database ontology approaches applicable to defining a therapeutic endpoint. The value of this strategy for drug discovery is evaluated, and perspectives for bioinformatics-driven hit identification are outlined.
Keywords: HIV; Influenza virus; RNAi; antiviral; bioinformatics; genome-wide screening; pathway analysis; siRNA; target identification.
Figures

References
-
- Coen DM, Richman DD. Antiviral Agents. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Vol. 1. Wolters Kluwer/Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 447–485.
-
- Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

