Comparative genomic analysis of the proteasome β5t subunit gene: implications for the origin and evolution of thymoproteasomes
- PMID: 21748441
- PMCID: PMC3805029
- DOI: 10.1007/s00251-011-0558-0
Comparative genomic analysis of the proteasome β5t subunit gene: implications for the origin and evolution of thymoproteasomes
Abstract
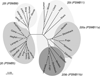
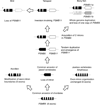
The thymoproteasome is a recently discovered, specialized form of 20S proteasomes expressed exclusively in the thymic cortex. Although the precise molecular mechanism by which the thymoproteasome exerts its function remains to be elucidated, accumulating evidence indicates that it plays a crucial role in positive selection of T cells. In the present study, we analyzed the evolution of the β5t subunit, a β-type catalytic subunit uniquely present in thymoproteasomes. The gene coding for the β5t subunit, designated PSMB11, was identified in the cartilaginous fish, the most divergent group of jawed vertebrates compared to the other jawed vertebrates, but not in jawless vertebrates or invertebrates. Interestingly, teleost fish have two copies of apparently functional PSMB11 genes, designated PSMB11a and PSMB11b, that encode β5t subunits with distinct amino acids in the S1 pocket. BLAST searches of genome databases suggest that birds such as chickens, turkey, and zebra finch lost the PSMB11 gene, and have neither thymoproteasomes nor immunoproteasomes. In mammals, reptiles, amphibians, and teleost fishes, the PSMB11 gene (the PSMB11a gene in teleost fish) is located next to the PSMB5 gene coding for the β5 subunit of the standard 20S proteasome, indicating that the PSMB11 gene arose by tandem duplication from the evolutionarily more ancient PSMB5 gene. The general absence of introns in PSMB11 and an unusual exon-intron structure of jawed vertebrate PSMB5 suggest that PSMB5 lost introns and duplicated in tandem in a common ancestor of jawed vertebrates, with PSMB5 subsequently gaining two introns and PSMB11 remaining intronless.
Figures



References
-
- Abdulla S, Beck S, Belich M, Jackson A, Nakamura T, Trowsdale J. Divergent intron arrangement in the MB1/LMP7 proteasome gene pair. Immunogenetics. 1996;44:254–258. - PubMed
-
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. - PubMed
-
- Bajoghli B, Guo P, Aghaallaei N, Hirano M, Strohmeier C, McCurley N, Bockman DE, Schorpp M, Cooper MD, Boehm T. A thymus candidate in lampreys. Nature. 2011;470:90–94. - PubMed
-
- Belich MP, Trowsdale J. Proteasome and class I antigen processing and presentation. Mol Biol Rep. 1995;21:53–56. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

