Population-averaged diffusion tensor imaging atlas of the Sprague Dawley rat brain
- PMID: 21749925
- PMCID: PMC3454512
- DOI: 10.1016/j.neuroimage.2011.06.063
Population-averaged diffusion tensor imaging atlas of the Sprague Dawley rat brain
Abstract
Rats are widely used in experimental neurobiological research, and rat brain atlases are important resources for identifying brain regions in the context of experimental microsurgery, tissue sampling, and neuroimaging, as well as comparison of findings across experiments. Currently, most available rat brain atlases are constructed from histological material derived from single specimens, and provide two-dimensional or three-dimensional (3D) outlines of diverse brain regions and fiber tracts. Important limitations of such atlases are that they represent individual specimens, and that finer details of tissue architecture are lacking. Access to more detailed 3D brain atlases representative of a population of animals is needed. Diffusion tensor imaging (DTI) is a unique neuroimaging modality that provides sensitive information about orientation structure in tissues, and is widely applied in basic and clinical neuroscience investigations. To facilitate analysis and assignment of location in rat brain neuroimaging investigations, we have developed a population-averaged three-dimensional DTI atlas of the normal adult Sprague Dawley rat brain. The atlas is constructed from high resolution ex vivo DTI images, which were nonlinearly warped into a population-averaged in vivo brain template. The atlas currently comprises a selection of manually delineated brain regions, the caudate-putamen complex, globus pallidus, entopeduncular nucleus, substantia nigra, external capsule, corpus callosum, internal capsule, cerebral peduncle, fimbria of the hippocampus, fornix, anterior commisure, optic tract, and stria terminalis. The atlas is freely distributed and potentially useful for several purposes, including automated and manual delineation of rat brain structural and functional imaging data.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
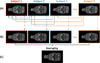

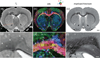

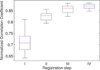
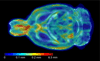

References
-
- Alexander DC, Pierpaoli C, Basser PJ, Gee JC. Spatial transformations of diffusion tensor magnetic resonance images. IEEE Trans Med Imaging. 2001;20(11):1131–1139. - PubMed
-
- Amaral D, Lavenex P. Hippocampal neuroanatomy. In: Andersen P, Morris R, Amaral D, Bliss T, O’Keefe J, editors. The Hippocampus. New York: Oxford University Press; 2007. Ch. 3.
-
- Antonsen BT, Jiang Y, Veraart J, Van Dongen Y, Qu H, Urbach Y, Raber K, Nguyen HP, Sijbers J, von Hörsten S, Johnson GA, Leergaard TB. Altered ex vivo diffusion mri in basal ganglia of transgenic huntington’s disease rats. 7th Forum of European Neuroscience Abstracts 0.46.2. 2010 Jul
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

