Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms
- PMID: 21749977
- PMCID: PMC3201869
- DOI: 10.1093/nar/gkr535
Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms
Abstract
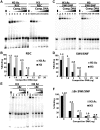
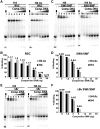
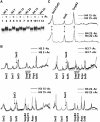
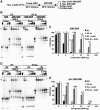
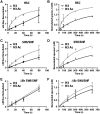
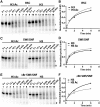
There is a close relationship between histone acetylation and ATP-dependent chromatin remodeling that is not fully understood. We show that acetylation of histone H3 tails affects SWI/SNF (mating type switching/ sucrose non fermenting) and RSC (remodels structure of chromatin) remodeling in several distinct ways. Acetylation of the histone H3 N-terminal tail facilitated recruitment and nucleosome mobilization by the ATP-dependent chromatin remodelers SWI/SNF and RSC. Tetra-acetylated H3, but not tetra-acetylated H4 tails, increased the affinity of RSC and SWI/SNF for nucleosomes while also changing the subunits of SWI/SNF that interact with the H3 tail. The enhanced recruitment of SWI/SNF due to H3 acetylation is bromodomain dependent, but is not further enhanced by additional bromodomains found in RSC. The combined effect of H3 acetylation and transcription activators is greater than either separately which suggests they act in parallel to recruit SWI/SNF. Besides enhancing recruitment, H3 acetylation increased nucleosome mobilization and H2A/H2B displacement by RSC and SWI/SNF in a bromodomain dependent manner and to a lesser extent enhanced ATP hydrolysis independent of bromodomains. H3 and H4 acetylation did not stimulate disassembly of adjacent nucleosomes in short arrays by SWI/SNF or RSC. These data illustrate how histone acetylation modulates RSC and SWI/SNF function, and provide a mechanistic insight into their collaborative efforts to remodel chromatin.
Figures

References
-
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. - PubMed
-
- Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A-H2B displacement. Nat. Struct. Mol. Biol. 2007;14:540–547. - PubMed
-
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell. 2004;14:667–673. - PubMed
-
- Chatterjee N, Sen P, Bartholomew B. In: Handbook of cell signalling. Bradshaw RA, Dennis EA, editors. Vol. 3. Oxford: Academic Press; 2009. pp. 2345–2356.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

