S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics
- PMID: 21749987
- PMCID: PMC3226405
- DOI: 10.1074/mcp.M111.009506
S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics
Abstract
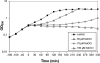
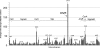
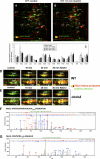
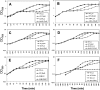
Protein S-thiolation is a post-translational thiol-modification that controls redox-sensing transcription factors and protects active site cysteine residues against irreversible oxidation. In Bacillus subtilis the MarR-type repressor OhrR was shown to sense organic hydroperoxides via formation of mixed disulfides with the redox buffer bacillithiol (Cys-GlcN-Malate, BSH), termed as S-bacillithiolation. Here we have studied changes in the transcriptome and redox proteome caused by the strong oxidant hypochloric acid in B. subtilis. The expression profile of NaOCl stress is indicative of disulfide stress as shown by the induction of the thiol- and oxidative stress-specific Spx, CtsR, and PerR regulons. Thiol redox proteomics identified only few cytoplasmic proteins with reversible thiol-oxidations in response to NaOCl stress that include GapA and MetE. Shotgun-liquid chromatography-tandem MS analyses revealed that GapA, Spx, and PerR are oxidized to intramolecular disulfides by NaOCl stress. Furthermore, we identified six S-bacillithiolated proteins in NaOCl-treated cells, including the OhrR repressor, two methionine synthases MetE and YxjG, the inorganic pyrophosphatase PpaC, the 3-D-phosphoglycerate dehydrogenase SerA, and the putative bacilliredoxin YphP. S-bacillithiolation of the OhrR repressor leads to up-regulation of the OhrA peroxiredoxin that confers together with BSH specific protection against NaOCl. S-bacillithiolation of MetE, YxjG, PpaC and SerA causes hypochlorite-induced methionine starvation as supported by the induction of the S-box regulon. The mechanism of S-glutathionylation of MetE has been described in Escherichia coli also leading to enzyme inactivation and methionine auxotrophy. In summary, our studies discover an important role of the bacillithiol redox buffer in protection against hypochloric acid by S-bacillithiolation of the redox-sensing regulator OhrR and of four enzymes of the methionine biosynthesis pathway.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

