The L6 domain tetraspanin Tm4sf4 regulates endocrine pancreas differentiation and directed cell migration
- PMID: 21750032
- PMCID: PMC3133913
- DOI: 10.1242/dev.058693
The L6 domain tetraspanin Tm4sf4 regulates endocrine pancreas differentiation and directed cell migration
Abstract
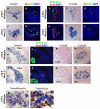
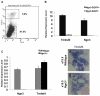
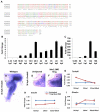
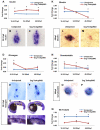
The homeodomain transcription factor Nkx2.2 is essential for pancreatic development and islet cell type differentiation. We have identified Tm4sf4, an L6 domain tetraspanin family member, as a transcriptional target of Nkx2.2 that is greatly upregulated during pancreas development in Nkx2.2(-/-) mice. Tetraspanins and L6 domain proteins recruit other membrane receptors to form active signaling centers that coordinate processes such as cell adhesion, migration and differentiation. In this study, we determined that Tm4sf4 is localized to the ductal epithelial compartment and is prominent in the Ngn3(+) islet progenitor cells. We also established that pancreatic tm4sf4 expression and regulation by Nkx2.2 is conserved during zebrafish development. Loss-of-function studies in zebrafish revealed that tm4sf4 inhibits α and β cell specification, but is necessary for ε cell fates. Thus, Tm4sf4 functional output opposes that of Nkx2.2. Further investigation of how Tm4sf4 functions at the cellular level in vitro showed that Tm4sf4 inhibits Rho-activated cell migration and actin organization in a ROCK-independent fashion. We propose that the primary role of Nkx2.2 is to inhibit Tm4sf4 in endocrine progenitor cells, allowing for delamination, migration and/or appropriate cell fate decisions. Identification of a role for Tm4sf4 during endocrine differentiation provides insight into islet progenitor cell behaviors and potential targetable regenerative mechanisms.
Figures
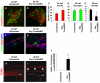
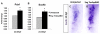
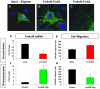
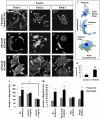

References
-
- Biemar F., Argenton F., Schmidtke R., Epperlein S., Peers B., Driever W. (2001). Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev. Biol. 230, 189-203 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

