Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity
- PMID: 21750106
- PMCID: PMC3202292
- DOI: 10.1101/gr.121541.111
Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity
Abstract
The human body contains thousands of unique cell types, each with specialized functions. Cell identity is governed in large part by gene transcription programs, which are determined by regulatory elements encoded in DNA. To identify regulatory elements active in seven cell lines representative of diverse human cell types, we used DNase-seq and FAIRE-seq (Formaldehyde Assisted Isolation of Regulatory Elements) to map "open chromatin." Over 870,000 DNaseI or FAIRE sites, which correspond tightly to nucleosome-depleted regions, were identified across the seven cell lines, covering nearly 9% of the genome. The combination of DNaseI and FAIRE is more effective than either assay alone in identifying likely regulatory elements, as judged by coincidence with transcription factor binding locations determined in the same cells. Open chromatin common to all seven cell types tended to be at or near transcription start sites and to be coincident with CTCF binding sites, while open chromatin sites found in only one cell type were typically located away from transcription start sites and contained DNA motifs recognized by regulators of cell-type identity. We show that open chromatin regions bound by CTCF are potent insulators. We identified clusters of open regulatory elements (COREs) that were physically near each other and whose appearance was coordinated among one or more cell types. Gene expression and RNA Pol II binding data support the hypothesis that COREs control gene activity required for the maintenance of cell-type identity. This publicly available atlas of regulatory elements may prove valuable in identifying noncoding DNA sequence variants that are causally linked to human disease.
Figures



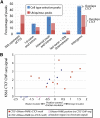
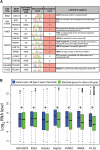

References
-
- Bell AC, West AG, Felsenfeld G 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
