NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation
- PMID: 21750686
- PMCID: PMC3131286
- DOI: 10.1371/journal.pgen.1002169
NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation
Abstract
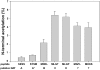
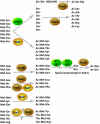
N-terminal acetylation (N-Ac) is a highly abundant eukaryotic protein modification. Proteomics revealed a significant increase in the occurrence of N-Ac from lower to higher eukaryotes, but evidence explaining the underlying molecular mechanism(s) is currently lacking. We first analysed protein N-termini and their acetylation degrees, suggesting that evolution of substrates is not a major cause for the evolutionary shift in N-Ac. Further, we investigated the presence of putative N-terminal acetyltransferases (NATs) in higher eukaryotes. The purified recombinant human and Drosophila homologues of a novel NAT candidate was subjected to in vitro peptide library acetylation assays. This provided evidence for its NAT activity targeting Met-Lys- and other Met-starting protein N-termini, and the enzyme was termed Naa60p and its activity NatF. Its in vivo activity was investigated by ectopically expressing human Naa60p in yeast followed by N-terminal COFRADIC analyses. hNaa60p acetylated distinct Met-starting yeast protein N-termini and increased general acetylation levels, thereby altering yeast in vivo acetylation patterns towards those of higher eukaryotes. Further, its activity in human cells was verified by overexpression and knockdown of hNAA60 followed by N-terminal COFRADIC. NatF's cellular impact was demonstrated in Drosophila cells where NAA60 knockdown induced chromosomal segregation defects. In summary, our study revealed a novel major protein modifier contributing to the evolution of N-Ac, redundancy among NATs, and an essential regulator of normal chromosome segregation. With the characterization of NatF, the co-translational N-Ac machinery appears complete since all the major substrate groups in eukaryotes are accounted for.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
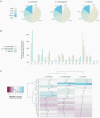


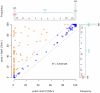
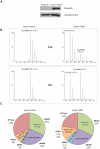
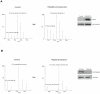

References
-
- Behnia R, Panic B, Whyte JR, Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol. 2004;6:405–413. - PubMed
-
- Caesar R, Blomberg A. The stress-induced Tfs1p requires NatB-mediated acetylation to inhibit carboxypeptidase Y and to regulate the protein kinase A pathway. J Biol Chem. 2004;279:38532–38543. - PubMed
-
- Setty SR, Strochlic TI, Tong AH, Boone C, Burd CG. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat Cell Biol. 2004;6:414–419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

