Dissecting the first transcriptional divergence during human embryonic development
- PMID: 21750961
- PMCID: PMC3285757
- DOI: 10.1007/s12015-011-9301-3
Dissecting the first transcriptional divergence during human embryonic development
Abstract
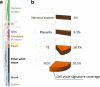
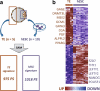
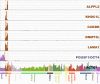
The trophoblast cell lineage is specified early at the blastocyst stage, leading to the emergence of the trophectoderm and the pluripotent cells of the inner cell mass. Using a double mRNA amplification technique and a comparison with transcriptome data on pluripotent stem cells, placenta, germinal and adult tissues, we report here some essential molecular features of the human mural trophectoderm. In addition to genes known for their role in placenta (CGA, PGF, ALPPL2 and ABCG2), human trophectoderm also strongly expressed Laminins, such as LAMA1, and the GAGE Cancer/Testis genes. The very high level of ABCG2 expression in trophectoderm, 7.9-fold higher than in placenta, suggests a major role of this gene in shielding the very early embryo from xenobiotics. Several genes, including CCKBR and DNMT3L, were specifically up-regulated only in trophectoderm, indicating that the trophoblast cell lineage shares with the germinal lineage a transient burst of DNMT3L expression. A trophectoderm core transcriptional regulatory circuitry formed by 13 tightly interconnected transcription factors (CEBPA, GATA2, GATA3, GCM1, KLF5, MAFK, MSX2, MXD1, PPARD, PPARG, PPP1R13L, TFAP2C and TP63), was found to be induced in trophectoderm and maintained in placenta. The induction of this network could be recapitulated in an in vitro trophoblast differentiation model.
Figures

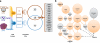
References
-
- Wong, C. C., Loewke, K. E., Bossert, N. L., et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nature Biotechnology, 28, 1115–1121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

