Human immunodeficiency virus rev-binding protein is essential for influenza a virus replication and promotes genome trafficking in late-stage infection
- PMID: 21752912
- PMCID: PMC3165756
- DOI: 10.1128/JVI.05064-11
Human immunodeficiency virus rev-binding protein is essential for influenza a virus replication and promotes genome trafficking in late-stage infection
Abstract
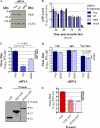
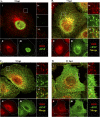
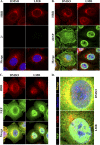
Influenza A virus uses cellular protein transport systems (e.g., CRM1-mediated nuclear export and Rab11-dependent recycling endosomes) for genome trafficking from the nucleus to the plasma membrane, where new virions are assembled. However, the detailed mechanisms of these events have not been completely resolved, and additional cellular factors are probably required. Here, we investigated the role of the cellular human immunodeficiency virus (HIV) Rev-binding protein (HRB), which interacts with influenza virus nuclear export protein (NEP), during the influenza virus life cycle. By using small interfering RNAs (siRNAs) and overexpression of a dominant negative HRB protein fragment, we show that cells lacking functional HRB have significantly reduced production of influenza virus progeny and that this defect results from impaired viral ribonucleoprotein (vRNP) delivery to the plasma membrane in late-stage infection. Since HRB colocalizes with influenza vRNPs early after their delivery to the cytoplasm, it may mediate a connection between the nucleocytoplasmic transport machinery and the endosomal system, thus facilitating the transfer of vRNPs from nuclear export to cytoplasmic trafficking complexes. We also found an association between NEP and HRB in the perinuclear region, suggesting that NEP may contribute to this process. Our results identify HRB as a second endosomal factor with a crucial role in influenza virus genome trafficking, suggest cooperation between unique endosomal compartments in the late steps of the influenza virus life cycle, and provide a common link between the cytoplasmic trafficking mechanisms of influenza virus and HIV.
Figures




Similar articles
-
Influenza virus-induced caspase-dependent enlargement of nuclear pores promotes nuclear export of viral ribonucleoprotein complexes.J Virol. 2015 Jun;89(11):6009-21. doi: 10.1128/JVI.03531-14. Epub 2015 Mar 25. J Virol. 2015. PMID: 25810542 Free PMC article.
-
Influenza B and C virus NEP (NS2) proteins possess nuclear export activities.J Virol. 2001 Aug;75(16):7375-83. doi: 10.1128/JVI.75.16.7375-7383.2001. J Virol. 2001. PMID: 11462009 Free PMC article.
-
Characteristics of nucleocytoplasmic transport of H1N1 influenza A virus nuclear export protein.J Virol. 2014 Jul;88(13):7455-63. doi: 10.1128/JVI.00257-14. Epub 2014 Apr 16. J Virol. 2014. PMID: 24741105 Free PMC article.
-
Influenza A virus RNA localisation and the interceding trafficking pathways of the host cell.PLoS Pathog. 2025 Apr 23;21(4):e1013090. doi: 10.1371/journal.ppat.1013090. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40267083 Free PMC article. Review.
-
Emerging roles for the influenza A virus nuclear export protein (NEP).PLoS Pathog. 2012;8(12):e1003019. doi: 10.1371/journal.ppat.1003019. Epub 2012 Dec 6. PLoS Pathog. 2012. PMID: 23236273 Free PMC article. Review.
Cited by
-
A Comprehensive Review on the Interaction Between the Host GTPase Rab11 and Influenza A Virus.Front Cell Dev Biol. 2019 Jan 9;6:176. doi: 10.3389/fcell.2018.00176. eCollection 2018. Front Cell Dev Biol. 2019. PMID: 30687703 Free PMC article. Review.
-
ITGB5 and AGFG1 variants are associated with severity of airway responsiveness.BMC Med Genet. 2013 Aug 28;14:86. doi: 10.1186/1471-2350-14-86. BMC Med Genet. 2013. PMID: 23984888 Free PMC article.
-
The influenza A virus PB2, PA, NP, and M segments play a pivotal role during genome packaging.J Virol. 2012 Jul;86(13):7043-51. doi: 10.1128/JVI.00662-12. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532680 Free PMC article.
-
The Feat of Packaging Eight Unique Genome Segments.Viruses. 2016 Jun 17;8(6):165. doi: 10.3390/v8060165. Viruses. 2016. PMID: 27322310 Free PMC article. Review.
-
Transport of the influenza virus genome from nucleus to nucleus.Viruses. 2013 Oct 2;5(10):2424-46. doi: 10.3390/v5102424. Viruses. 2013. PMID: 24104053 Free PMC article. Review.
References
-
- Bogerd H. P., Fridell R. A., Madore S., Cullen B. R. 1995. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell 82:485–494 - PubMed
-
- Bowzard J. B., Cheng D., Peng J., Kahn R. A. 2007. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J. Biol. Chem. 282:17568–17580 - PubMed
-
- Bruce E. A., et al. 2009. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4- and VPS28-independent pathway. Virology 390:268–278 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

