The HIV-1 Tat protein has a versatile role in activating viral transcription
- PMID: 21752913
- PMCID: PMC3165771
- DOI: 10.1128/JVI.00650-11
The HIV-1 Tat protein has a versatile role in activating viral transcription
Abstract
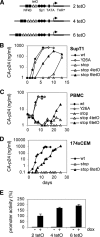
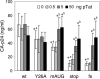
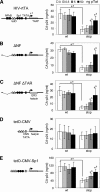
It is generally acknowledged that the Tat protein has a pivotal role in HIV-1 replication because it stimulates transcription from the viral long terminal repeat (LTR) promoter by binding to the TAR hairpin in the nascent RNA transcript. However, a multitude of additional Tat functions have been suggested. The importance of these functions is difficult to assess in replication studies with Tat-mutated HIV-1 variants because of the dominant negative effect on viral gene expression. We therefore used an HIV-1 construct that does not depend on the Tat-TAR interaction for transcription to reevaluate whether or not Tat has a second essential function in HIV-1 replication. This HIV-rtTA variant uses the incorporated Tet-On gene expression system for activation of transcription and replicates efficiently upon complete TAR deletion. Here we demonstrated that Tat inactivation does nevertheless severely inhibit replication. Upon long-term culturing, the Tat-minus HIV-rtTA variant acquired mutations in the U3 region that improved promoter activity and reestablished replication. We showed that in the absence of a functional TAR, Tat remains important for viral transcription via Sp1 sequence elements in the U3 promoter region. Substitution of these U3 sequences with nonrelated promoter elements created a virus that replicates efficiently without Tat in SupT1 T cells. These results indicate that Tat has a versatile role in transcription via TAR and U3 elements. The results also imply that Tat has no other essential function in viral replication in cultured T cells.
Figures




Similar articles
-
Tat has a dual role in simian immunodeficiency virus transcription.J Gen Virol. 2012 Oct;93(Pt 10):2279-2289. doi: 10.1099/vir.0.044511-0. Epub 2012 Jul 18. J Gen Virol. 2012. PMID: 22815271
-
Semen Exosomes Promote Transcriptional Silencing of HIV-1 by Disrupting NF-κB/Sp1/Tat Circuitry.J Virol. 2018 Oct 12;92(21):e00731-18. doi: 10.1128/JVI.00731-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111566 Free PMC article.
-
Tat functions to stimulate the elongation properties of transcription complexes paused by the duplicated TAR RNA element of human immunodeficiency virus 2.J Mol Biol. 1995 Dec 1;254(3):350-63. doi: 10.1006/jmbi.1995.0622. J Mol Biol. 1995. PMID: 7490754
-
Tackling Tat.J Mol Biol. 1999 Oct 22;293(2):235-54. doi: 10.1006/jmbi.1999.3060. J Mol Biol. 1999. PMID: 10550206 Review.
-
Face-time with TAR: Portraits of an HIV-1 RNA with diverse modes of effector recognition relevant for drug discovery.J Biol Chem. 2019 Jun 14;294(24):9326-9341. doi: 10.1074/jbc.REV119.006860. Epub 2019 May 12. J Biol Chem. 2019. PMID: 31080171 Free PMC article. Review.
Cited by
-
Drosophila as a Model for Infectious Diseases.Int J Mol Sci. 2021 Mar 8;22(5):2724. doi: 10.3390/ijms22052724. Int J Mol Sci. 2021. PMID: 33800390 Free PMC article. Review.
-
HIV-1 Tat Upregulates the Receptor for Advanced Glycation End Products and Superoxide Dismutase-2 in the Heart of Transgenic Mice.Viruses. 2022 Oct 4;14(10):2191. doi: 10.3390/v14102191. Viruses. 2022. PMID: 36298745 Free PMC article.
-
IFI16 Targets the Transcription Factor Sp1 to Suppress HIV-1 Transcription and Latency Reactivation.Cell Host Microbe. 2019 Jun 12;25(6):858-872.e13. doi: 10.1016/j.chom.2019.05.002. Epub 2019 Jun 4. Cell Host Microbe. 2019. PMID: 31175045 Free PMC article.
-
Control of viral infections by epigenetic-targeted therapy.Clin Epigenetics. 2019 Mar 27;11(1):55. doi: 10.1186/s13148-019-0654-9. Clin Epigenetics. 2019. PMID: 30917875 Free PMC article. Review.
-
New insights into pathogenesis point to HIV-1 Tat as a key vaccine target.Arch Virol. 2021 Nov;166(11):2955-2974. doi: 10.1007/s00705-021-05158-z. Epub 2021 Aug 14. Arch Virol. 2021. PMID: 34390393 Free PMC article. Review.
References
-
- Apolloni A., Meredith L. W., Suhrbier A., Kiernan R., Harrich D. 2007. The HIV-1 Tat protein stimulates reverse transcription in vitro. Curr. HIV Res. 5:473–483 - PubMed
-
- Bannwarth S., Gatignol A. 2005. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr. HIV Res. 3:61–71 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

