The structure of barmah forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions
- PMID: 21752915
- PMCID: PMC3165765
- DOI: 10.1128/JVI.05015-11
The structure of barmah forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions
Abstract
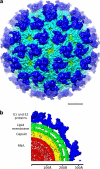
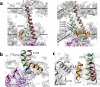
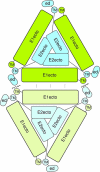
Barmah Forest virus (BFV) is a mosquito-borne alphavirus that infects humans. A 6-Å-resolution cryo-electron microscopy three-dimensional structure of BFV exhibits a typical alphavirus organization, with RNA-containing nucleocapsid surrounded by a bilipid membrane anchored with the surface proteins E1 and E2. The map allows details of the transmembrane regions of E1 and E2 to be seen. The C-terminal end of the E2 transmembrane helix binds to the capsid protein. Following the E2 transmembrane helix, a short α-helical endodomain lies on the inner surface of the lipid envelope. The E2 endodomain interacts with E1 transmembrane helix from a neighboring E1-E2 trimeric spike, thereby acting as a spacer and a linker between spikes. In agreement with previous mutagenesis studies, the endodomain plays an important role in recruiting other E1-E2 spikes to the budding site during virus assembly. The E2 endodomain may thus serve as a target for antiviral drug design.
Figures


References
-
- Caspar D. L., Klug A. 1962. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27:1–24 - PubMed
-
- Charollais J., Van Der Goot F. G. 2009. Palmitoylation of membrane proteins (review). Mol. Membr. Biol. 26:55–66 - PubMed
-
- Choi H. K., Lu G., Lee S., Wengler G., Rossmann M. G. 1997. Structure of Semliki Forest virus core protein. Proteins 27:345–359 - PubMed
-
- Combet C., Blanchet C., Geourjon C., Deleage G. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147–150 - PubMed
-
- Dalgarno L., et al. 1984. Characterization of Barmah forest virus: an alphavirus with some unusual properties. Virology 133:416–426 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

